Protein folding studied by single-molecule FRET
- PMID: 18221865
- PMCID: PMC2323684
- DOI: 10.1016/j.sbi.2007.12.003
Protein folding studied by single-molecule FRET
Abstract
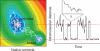
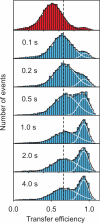
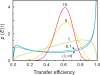
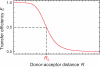
A complete understanding of a protein-folding mechanism requires description of the distribution of microscopic pathways that connect the folded and unfolded states. This distribution can, in principle, be described by computer simulations and theoretical models of protein folding, but is hidden in conventional experiments on large ensembles of molecules because only average properties are measured. A long-term goal of single-molecule fluorescence studies is to time-resolve the structural events as individual molecules make transitions between folded and unfolded states. Although such studies are still in their infancy, the work till now shows great promise and has already produced novel and important information on current issues in protein folding that has been impossible or difficult to obtain from ensemble measurements.
Figures

References
-
- Fisher TE, Oberhauser AF, Carrion-Vazquez M, Marszalek PE, Fernandez JM. The study of protein mechanics with the atomic force microscope. Trends in Biochemical Sciences. 1999;24:379–384. - PubMed
-
- Zhuang X, Rief M. Single-molecule folding. Curr Opin Struct Biol. 2003;13:88–97. - PubMed
-
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. - PubMed
-
- Forman JR, Clarke J. Mechanical unfolding of proteins: insights into biology, structure and folding. Curr Opin Struct Biol. 2007;17:58–66. - PubMed
-
- Hummer G, Szabo A. Free energy surfaces from single-molecule force spectroscopy. Accounts Chem Res. 2005;38:504–513. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

