Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli
- PMID: 18222925
- PMCID: PMC2650629
- DOI: 10.1093/dnares/dsm033
Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli
Abstract
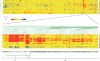
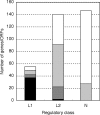
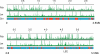
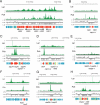
Enterohemorrhagic Escherichia coli is an emerging pathogen that causes diarrhea and hemolytic uremic syndrome. Much of the genomic information that affects virulence is acquired by horizontal transfer. Genes necessary for attaching and effacing lesions are located in the locus for enterocyte effacement (LEE) pathogenicity island. LEE gene transcription is positively regulated by Ler, which is also encoded by the LEE, and by Pch regulators, which are encoded at other loci. Here we identified genes whose transcription profiles were similar to those of the LEE genes, by comparing the effects of altering ler and pch transcript levels. We assigned these genes into two classes, according to their transcription profiles. By determining the binding profiles for Ler and Pch, we showed that both were involved in regulating one class of genes, but only Pch was involved in regulating the other class. Binding sites were found in the coding region as well as the promoter region of regulated genes, which include genes common to K12 strains as well as 0157-specific genes, suggesting that both act as a global regulator. These results indicate that Ler and Pch orchestrate the transcription of virulence genes, which are captured by horizontal transfer and scattered throughout the chromosome.
Figures

References
-
- Gal-Mor O., Finlay B. B. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. - PubMed
-
- Kaper J. B., Nataro J. P., Mobley H. L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. - PubMed
-
- Hayashi T., Makino K., Ohnishi M., et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. - PubMed
-
- Frankel G., Phillips A. D., Rosenshine I., et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 1998;30:911–921. - PubMed
-
- Hayward R. D., Leong J. M., Koronakis V., et al. Exploiting pathogenic Escherichia coli to model transmembrane receptor signalling. Nat. Rev. Microbiol. 2006;4:358–370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

