EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol
- PMID: 18223037
- PMCID: PMC2254931
- DOI: 10.1105/tpc.107.055178
EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol
Abstract
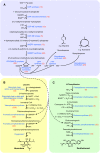
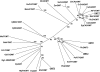
The glandular trichomes (lupulin glands) of hop (Humulus lupulus) synthesize essential oils and terpenophenolic resins, including the bioactive prenylflavonoid xanthohumol. To dissect the biosynthetic processes occurring in lupulin glands, we sequenced 10,581 ESTs from four trichome-derived cDNA libraries. ESTs representing enzymes of terpenoid biosynthesis, including all of the steps of the methyl 4-erythritol phosphate pathway, were abundant in the EST data set, as were ESTs for the known type III polyketide synthases of bitter acid and xanthohumol biosynthesis. The xanthohumol biosynthetic pathway involves a key O-methylation step. Four S-adenosyl-l-methionine-dependent O-methyltransferases (OMTs) with similarity to known flavonoid-methylating enzymes were present in the EST data set. OMT1, which was the most highly expressed OMT based on EST abundance and RT-PCR analysis, performs the final reaction in xanthohumol biosynthesis by methylating desmethylxanthohumol to form xanthohumol. OMT2 accepted a broad range of substrates, including desmethylxanthohumol, but did not form xanthohumol. Mass spectrometry and proton nuclear magnetic resonance analysis showed it methylated xanthohumol to 4-O-methylxanthohumol, which is not known from hop. OMT3 was inactive with all substrates tested. The lupulin gland-specific EST data set expands the genomic resources for H. lupulus and provides further insight into the metabolic specialization of glandular trichomes.
Figures



References
-
- Audic, S., and Claverie, J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7 986–995. - PubMed
-
- Biendl, M. (2003 2002). Research on the xanthohumol content of hops. Hopfenrundschau International 2002/2003 72–75.
-
- Bonaldo, M.F., Lennon, G., and Soares, M.B. (1996). Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 6 791–806. - PubMed
-
- Burga, L., Wellmann, F., Lukacin, R., Witte, S., Schwab, W., Schroder, J., and Matern, U. (2005). Unusual pseudosubstrate specificity of a novel 3,5-dimethoxyphenol O-methyltransferase cloned from Ruta graveolens L. Arch. Biochem. Biophys. 440 54–64. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

