Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli
- PMID: 18223071
- PMCID: PMC2293195
- DOI: 10.1128/JB.01353-07
Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli
Abstract
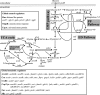
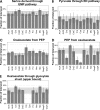
Although a whole arsenal of mechanisms are potentially involved in metabolic regulation, it is largely uncertain when, under which conditions, and to which extent a particular mechanism actually controls network fluxes and thus cellular physiology. Based on (13)C flux analysis of Escherichia coli mutants, we elucidated the relevance of global transcriptional regulation by ArcA, ArcB, Cra, CreB, CreC, Crp, Cya, Fnr, Hns, Mlc, OmpR, and UspA on aerobic glucose catabolism in glucose-limited chemostat cultures at a growth rate of 0.1 h(-1). The by far most relevant control mechanism was cyclic AMP (cAMP)-dependent catabolite repression as the inducer of the phosphoenolpyruvate (PEP)-glyoxylate cycle and thus low tricarboxylic acid cycle fluxes. While all other mutants and the reference E. coli strain exhibited high glyoxylate shunt and PEP carboxykinase fluxes, and thus high PEP-glyoxylate cycle flux, this cycle was essentially abolished in both the Crp and Cya mutants, which lack the cAMP-cAMP receptor protein complex. Most other mutations were phenotypically silent, and only the Cra and Hns mutants exhibited slightly altered flux distributions through PEP carboxykinase and the tricarboxylic acid cycle, respectively. The Cra effect on PEP carboxykinase was probably the consequence of a specific control mechanism, while the Hns effect appears to be unspecific. For central metabolism, the available data thus suggest that a single transcriptional regulation process exerts the dominant control under a given condition and this control is highly specific for a single pathway or cycle within the network.
Figures


Similar articles
-
Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli.J Bacteriol. 2005 May;187(9):3171-9. doi: 10.1128/JB.187.9.3171-3179.2005. J Bacteriol. 2005. PMID: 15838044 Free PMC article.
-
Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli.Mol Syst Biol. 2011 Mar 29;7:477. doi: 10.1038/msb.2011.9. Mol Syst Biol. 2011. PMID: 21451587 Free PMC article.
-
Catabolic regulation analysis of Escherichia coli and its crp, mlc, mgsA, pgi and ptsG mutants.Microb Cell Fact. 2011 Aug 11;10:67. doi: 10.1186/1475-2859-10-67. Microb Cell Fact. 2011. PMID: 21831320 Free PMC article.
-
Inactivation of the PTS as a Strategy to Engineer the Production of Aromatic Metabolites in Escherichia coli.J Mol Microbiol Biotechnol. 2015;25(2-3):195-208. doi: 10.1159/000380854. Epub 2015 Jul 9. J Mol Microbiol Biotechnol. 2015. PMID: 26159079 Review.
-
Alteration of the biochemical valves in the central metabolism of Escherichia coli.Ann N Y Acad Sci. 1994 Nov 30;745:21-34. doi: 10.1111/j.1749-6632.1994.tb44361.x. Ann N Y Acad Sci. 1994. PMID: 7832509 Review.
Cited by
-
Metabolic flux configuration determination using information entropy.PLoS One. 2020 Dec 4;15(12):e0243067. doi: 10.1371/journal.pone.0243067. eCollection 2020. PLoS One. 2020. PMID: 33275628 Free PMC article.
-
Controlled burn: interconnections between energy-spilling pathways and metabolic signaling in bacteria.J Bacteriol. 2025 May 22;207(5):e0054224. doi: 10.1128/jb.00542-24. Epub 2025 Mar 31. J Bacteriol. 2025. PMID: 40162839 Free PMC article. Review.
-
Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase.BMC Syst Biol. 2010 Dec 1;4:166. doi: 10.1186/1752-0509-4-166. BMC Syst Biol. 2010. PMID: 21122111 Free PMC article.
-
A Metabolic Widget Adjusts the Phosphoenolpyruvate-Dependent Fructose Influx in Pseudomonas putida.mSystems. 2016 Dec 6;1(6):e00154-16. doi: 10.1128/mSystems.00154-16. eCollection 2016 Nov-Dec. mSystems. 2016. PMID: 27933319 Free PMC article.
-
Metabolic and regulatory rearrangements underlying efficient D-xylose utilization in engineered Pseudomonas putida S12.J Biol Chem. 2012 Apr 27;287(18):14606-14. doi: 10.1074/jbc.M111.337501. Epub 2012 Mar 13. J Biol Chem. 2012. PMID: 22416130 Free PMC article.
References
-
- Al Zaid Siddiquee, K., M. J. Arauzo-Bravo, and K. Shimizu. 2004. Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 63407-417. - PubMed
-
- Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 247-17. - PubMed
-
- Avison, M. B., R. E. Horton, T. R. Walsh, and P. M. Bennett. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J. Biol. Chem. 27626955-26961. - PubMed
-
- Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209141-148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

