Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration
- PMID: 18223198
- PMCID: PMC2900863
- DOI: 10.1093/hmg/ddn023
Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration
Abstract
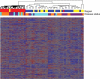
Frontotemporal lobar degeneration is a fatal neurodegenerative disease that results in progressive decline in behavior, executive function and sometimes language. Disease mechanisms remain poorly understood. Recently, however, the DNA- and RNA-binding protein TDP-43 has been identified as the major protein present in the hallmark inclusion bodies of frontotemporal lobar degeneration with ubiquitinated inclusions (FTLD-U), suggesting a role for transcriptional dysregulation in FTLD-U pathophysiology. Using the Affymetrix U133A microarray platform, we profiled global gene expression in both histopathologically affected and unaffected areas of human FTLD-U brains. We then characterized differential gene expression with biological pathway analyses, cluster and principal component analyses, and subgroup analyses based on brain region and progranulin (GRN) gene status. Comparing 17 FTLD-U brains to 11 controls, we identified 414 upregulated and 210 downregulated genes in frontal cortex (P-value < 0.001). Moreover, cluster and principal component analyses revealed that samples with mutations or possibly pathogenic variations in the GRN gene (GRN+, 7/17) had an expression signature that was distinct from both normal controls and FTLD-U samples lacking GRN gene variations (GRN-, 10/17). Within the subgroup of GRN+ FTLD-U, we found >1300 dysregulated genes in frontal cortex (P-value < 0.001), many participating in pathways uniquely dysregulated in the GRN+ cases. Our findings demonstrate a distinct molecular phenotype for GRN+ FTLD-U, not readily apparent on clinical or histopathological examination, suggesting distinct pathophysiological mechanisms for GRN+ and GRN- subtypes of FTLD-U. In addition, these data from a large number of human brains provide a valuable resource for future testing of disease hypotheses.
Figures
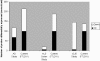


References
-
- Ratnavalli E., Brayne C., Dawson K., Hodges J.R. The prevalence of frontotemporal dementia. Neurology. 2002;58:1615–1621. - PubMed
-
- McKhann G.M., Albert M.S., Grossman M., Miller B., Dickson D., Trojanowski J.Q. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch. Neurol. 2001;58:1803–1809. - PubMed
-
- Neary D., Snowden J.S., Gustafson L., Passant U., Stuss D., Black S., Freedman M., Kertesz A., Robert P.H., Albert M., et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

