Genomic sequence is highly predictive of local nucleosome depletion
- PMID: 18225943
- PMCID: PMC2211532
- DOI: 10.1371/journal.pcbi.0040013
Genomic sequence is highly predictive of local nucleosome depletion
Abstract
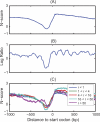
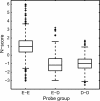
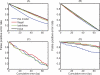
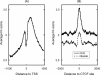
The regulation of DNA accessibility through nucleosome positioning is important for transcription control. Computational models have been developed to predict genome-wide nucleosome positions from DNA sequences, but these models consider only nucleosome sequences, which may have limited their power. We developed a statistical multi-resolution approach to identify a sequence signature, called the N-score, that distinguishes nucleosome binding DNA from non-nucleosome DNA. This new approach has significantly improved the prediction accuracy. The sequence information is highly predictive for local nucleosome enrichment or depletion, whereas predictions of the exact positions are only modestly more accurate than a null model, suggesting the importance of other regulatory factors in fine-tuning the nucleosome positions. The N-score in promoter regions is negatively correlated with gene expression levels. Regulatory elements are enriched in low N-score regions. While our model is derived from yeast data, the N-score pattern computed from this model agrees well with recent high-resolution protein-binding data in human.
Conflict of interest statement
Figures



References
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
-
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. - PubMed
-
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

