Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein
- PMID: 18225958
- PMCID: PMC2213710
- DOI: 10.1371/journal.pgen.0040021
Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein
Abstract
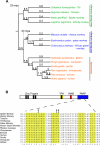
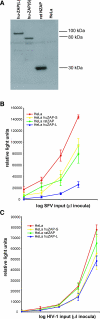
Intrinsic immunity relies on specific recognition of viral epitopes to mount a cell-autonomous defense against viral infections. Viral recognition determinants in intrinsic immunity genes are expected to evolve rapidly as host genes adapt to changing viruses, resulting in a signature of adaptive evolution. Zinc-finger antiviral protein (ZAP) from rats was discovered to be an intrinsic immunity gene that can restrict murine leukemia virus, and certain alphaviruses and filoviruses. Here, we used an approach combining molecular evolution and cellular infectivity assays to address whether ZAP also acts as a restriction factor in primates, and to pinpoint which protein domains may directly interact with the virus. We find that ZAP has evolved under positive selection throughout primate evolution. Recurrent positive selection is only found in the poly(ADP-ribose) polymerase (PARP)-like domain present in a longer human ZAP isoform. This PARP-like domain was not present in the previously identified and tested rat ZAP gene. Using infectivity assays, we found that the longer isoform of ZAP that contains the PARP-like domain is a stronger suppressor of murine leukemia virus expression and Semliki forest virus infection. Our study thus finds that human ZAP encodes a potent antiviral activity against alphaviruses. The striking congruence between our evolutionary predictions and cellular infectivity assays strongly validates such a combined approach to study intrinsic immunity genes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

References
-
- Bieniasz PD. Intrinsic immunity: A front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. - PubMed
-
- Goff SP. Retrovirus restriction factors. Mol Cell. 2004;16:849–859. - PubMed
-
- Goff SP. Genetic control of retrovirus susceptibility in mammalian cells. Annu Rev Genet. 2004;38:61–85. - PubMed
-
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:e275. doi: 10.1371/journal.pbio.0020275. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

