Evolution of a family of metazoan active-site-serine enzymes from penicillin-binding proteins: a novel facet of the bacterial legacy
- PMID: 18226203
- PMCID: PMC2266909
- DOI: 10.1186/1471-2148-8-26
Evolution of a family of metazoan active-site-serine enzymes from penicillin-binding proteins: a novel facet of the bacterial legacy
Abstract
Background: Bacterial penicillin-binding proteins and beta-lactamases (PBP-betaLs) constitute a large family of serine proteases that perform essential functions in the synthesis and maintenance of peptidoglycan. Intriguingly, genes encoding PBP-betaL homologs occur in many metazoan genomes including humans. The emerging role of LACTB, a mammalian mitochondrial PBP-betaL homolog, in metabolic signaling prompted us to investigate the evolutionary history of metazoan PBP-betaL proteins.
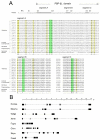
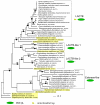
Results: Metazoan PBP-betaL homologs including LACTB share unique structural features with bacterial class B low molecular weight penicillin-binding proteins. The amino acid residues necessary for enzymatic activity in bacterial PBP-betaL proteins, including the catalytic serine residue, are conserved in all metazoan homologs. Phylogenetic analysis indicated that metazoan PBP-betaL homologs comprise four alloparalogus protein lineages that derive from alpha-proteobacteria.
Conclusion: While most components of the peptidoglycan synthesis machinery were dumped by early eukaryotes, a few PBP-betaL proteins were conserved and are found in metazoans including humans. Metazoan PBP-betaL homologs are active-site-serine enzymes that probably have distinct functions in the metabolic circuitry. We hypothesize that PBP-betaL proteins in the early eukaryotic cell enabled the degradation of peptidoglycan from ingested bacteria, thereby maximizing the yield of nutrients and streamlining the cell for effective phagocytotic feeding.
Figures

References
-
- Goffin C, Ghuysen J-M. Biochemistry and Comparative genomics of SxxK Superfamily Acyltransferases Offer a Clue to the Mycobacterial Paradox: Presence of Penicillin-Susceptible Target Proteins versus Lack of Efficiency of Penicillin as Therapeutic Agent. Microbiol Mol Biol Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

