Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages
- PMID: 18226223
- PMCID: PMC2267772
- DOI: 10.1186/1741-7007-6-6
Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages
Abstract
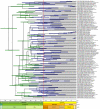
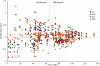
Background: Determining an absolute timescale for avian evolutionary history has proven contentious. The two sources of information available, paleontological data and inference from extant molecular genetic sequences (colloquially, 'rocks' and 'clocks'), have appeared irreconcilable; the fossil record supports a Cenozoic origin for most modern lineages, whereas molecular genetic estimates suggest that these same lineages originated deep within the Cretaceous and survived the K-Pg (Cretaceous-Paleogene; formerly Cretaceous-Tertiary or K-T) mass-extinction event. These two sources of data therefore appear to support fundamentally different models of avian evolution. The paradox has been speculated to reflect deficiencies in the fossil record, unrecognized biases in the treatment of genetic data or both. Here we attempt to explore uncertainty and limit bias entering into molecular divergence time estimates through: (i) improved taxon (n = 135) and character (n = 4594 bp mtDNA) sampling; (ii) inclusion of multiple cladistically tested internal fossil calibration points (n = 18); (iii) correction for lineage-specific rate heterogeneity using a variety of methods (n = 5); (iv) accommodation of uncertainty in tree topology; and (v) testing for possible effects of episodic evolution.
Results: The various 'relaxed clock' methods all indicate that the major (basal) lineages of modern birds originated deep within the Cretaceous, although temporal intraordinal diversification patterns differ across methods. We find that topological uncertainty had a systematic but minor influence on date estimates for the origins of major clades, and Bayesian analyses assuming fixed topologies deliver similar results to analyses with unconstrained topologies. We also find that, contrary to expectation, rates of substitution are not autocorrelated across the tree in an ancestor-descendent fashion. Finally, we find no signature of episodic molecular evolution related to either speciation events or the K-Pg boundary that could systematically mislead inferences from genetic data.
Conclusion: The 'rock-clock' gap has been interpreted by some to be a result of the vagaries of molecular genetic divergence time estimates. However, despite measures to explore different forms of uncertainty in several key parameters, we fail to reconcile molecular genetic divergence time estimates with dates taken from the fossil record; instead, we find strong support for an ancient origin of modern bird lineages, with many extant orders and families arising in the mid-Cretaceous, consistent with previous molecular estimates. Although there is ample room for improvement on both sides of the 'rock-clock' divide (e.g. accounting for 'ghost' lineages in the fossil record and developing more realistic models of rate evolution for molecular genetic sequences), the consistent and conspicuous disagreement between these two sources of data more likely reflects a genuine difference between estimated ages of (i) stem-group origins and (ii) crown-group morphological diversifications, respectively. Further progress on this problem will benefit from greater communication between paleontologists and molecular phylogeneticists in accounting for error in avian lineage age estimates.
Figures




References
-
- Archibald JD, Deutschman DH. Quantitative analysis of the timing of the origin and diversification of extant placental orders. J Mamm Evol. 2001;8:107–124. doi: 10.1023/A:1011317930838. - DOI
-
- van Tuinen M, Stidham TA, Hadly EA. Tempo and mode of modern bird evolution observed with large-scale taxonomic sampling. Hist Biol. 2006;18:205–221. doi: 10.1080/08912960600641174. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

