Estimating large-scale signaling networks through nested effect models with intervention effects from microarray data
- PMID: 18227117
- PMCID: PMC2579711
- DOI: 10.1093/bioinformatics/btm634
Estimating large-scale signaling networks through nested effect models with intervention effects from microarray data
Abstract
Motivation: Targeted interventions using RNA interference in combination with the measurement of secondary effects with DNA microarrays can be used to computationally reverse engineer features of upstream non-transcriptional signaling cascades based on the nested structure of effects.
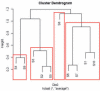
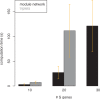
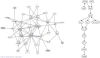
Results: We extend previous work by Markowetz et al., who proposed a statistical framework to score different network hypotheses. Our extensions go in several directions: we show how prior assumptions on the network structure can be incorporated into the scoring scheme by defining appropriate prior distributions on the network structure as well as on hyperparameters. An approach called module networks is introduced to scale up the original approach, which is limited to around 5 genes, to infer large-scale networks of more than 30 genes. Instead of the data discretization step needed in the original framework, we propose the usage of a beta-uniform mixture distribution on the P-value profile, resulting from differential gene expression calculation, to quantify effects. Extensive simulations on artificial data and application of our module network approach to infer the signaling network between 13 genes in the ER-alpha pathway in human MCF-7 breast cancer cells show that our approach gives sensible results. Using a bootstrapping and a jackknife approach, this reconstruction is found to be statistically stable.
Availability: The proposed method is available within the Bioconductor R-package nem.
Figures





References
-
- Aho A, et al. The transitive reduction of a directed graph. SIAM J. Comput. 1972;1:131–137.
-
- Boutros M, et al. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell. 2002;3:711–722. - PubMed
-
- Dempster A, et al. Maximum likelihood from incomplete data via the em algorithm. J. Royal Statistical Soc., Series B. 1977;39:1–38.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

