Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex
- PMID: 18227152
- PMCID: PMC2268434
- DOI: 10.1128/MCB.01886-07
Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex
Abstract
RNA editing in Trypanosoma brucei is posttranscriptional uridylate removal/addition, generally at vast numbers of pre-mRNA sites, but to date, only single editing cycles have been examined in vitro. We here demonstrate achieving sequential cycles of U deletion in vitro, with editing products confirmed by sequence analysis. Notably, the subsequent editing cycle is much more efficient and occurs far more rapidly than single editing cycles; plus, it has different recognition requirements. This indicates that the editing complex acts in a concerted manner and does not dissociate from the RNA substrate between these cycles. Furthermore, the multicycle substrate exhibits editing that is unexpected from a strictly 3'-to-5' progression, reminiscent of the unexpected editing that has been shown to occur frequently in T. brucei mRNAs edited in vivo. This unexpected editing is most likely due to alternate mRNA:guide RNA (gRNA) alignment forming a hyphenated anchor; its having only a 2-bp proximal duplex helps explain the prevalence of unexpected editing in vivo. Such unexpected editing was not previously reported in vitro, presumably because the common use of artificially tight mRNA:gRNA base pairing precludes alternate alignments. The multicycle editing and unexpected editing presented in this paper bring in vitro reactions closer to reproducing the in vivo editing process.
Figures
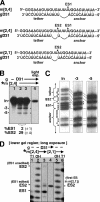
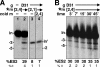


References
-
- Abraham, J. M., J. E. Feagin, and K. Stuart. 1988. Characterization of cytochrome c oxidase III transcripts that are edited only in the 3′ region. Cell 55267-272. - PubMed
-
- Benne, R., J. Van den Burg, J. P. Brakenhoff, P. Sloof, J. H. Van Boom, and M. C. Tromp. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46819-826. - PubMed
-
- Bhat, G. J., D. J. Koslowsky, J. E. Feagin, B. L. Smiley, and K. Stuart. 1990. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell 61885-894. - PubMed
-
- Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60189-198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
