Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene
- PMID: 18227185
- PMCID: PMC2292522
- DOI: 10.1128/AAC.01451-07
Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene
Abstract
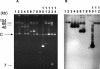
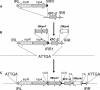
Genetic structures surrounding the carbapenem-hydrolyzing Ambler class A bla KPC gene were characterized in several KPC-positive Klebsiella pneumoniae and Pseudomonas aeruginosa strains isolated from the United States, Colombia, and Greece. The bla KPC genes were associated in all cases with transposon-related structures. In the K. pneumoniae YC isolate from the United States, the beta-lactamase bla KPC-2 gene was located on a novel Tn3-based transposon, Tn4401. Tn4401 was 10 kb in size, was delimited by two 39-bp imperfect inverted repeat sequences, and harbored, in addition to the beta-lactamase bla KPC-2 gene, a transposase gene, a resolvase gene, and two novel insertion sequences, ISKpn6 and ISKpn7. Tn4401 has been identified in all isolates. However, two isoforms of this transposon were found: Tn4401a was found in K. pneumoniae YC and K. pneumoniae GR from the United States and Greece, respectively, and differed by a 100-bp deletion, located just upstream of the bla KPC-2 gene, compared to the sequence of Tn4401b, which was found in the Colombian isolates. In all isolates tested, Tn4401 was flanked by a 5-bp target site duplication, the signature of a recent transposition event, and was inserted in different open reading frames located on plasmids that varied in size and nature. Tn4401 is likely at the origin of carbapenem-hydrolyzing beta-lactamase KPC mobilization to plasmids and its further insertion into various-sized plasmids identified in nonclonally related K. pneumoniae and P. aeruginosa isolates.
Figures



References
-
- Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55-60. - PubMed
-
- Bratu, S., S. Brooks, S. Burney, S. Kochar, J. Gupta, D. Landman, and J. Quale. 2007. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin. Infect. Dis. 44:972-975. - PubMed
-
- Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

