Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae
- PMID: 18230162
- PMCID: PMC2275737
- DOI: 10.1186/1471-2164-9-55
Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae
Abstract
Background: Proteins secreted by bacteria play an important role in infection of eukaryotic hosts. Rhizobia infect the roots of leguminous plants and establish a mutually beneficial symbiosis. Proteins secreted during the infection process by some rhizobial strains can influence infection and modify the plant defence signalling pathways. The aim of this study was to systematically analyse protein secretion in the recently sequenced strain Rhizobium leguminosarum bv. viciae 3841.
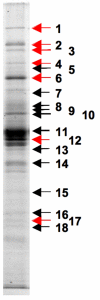
Results: Similarity searches using defined protein secretion systems from other Gram-negative bacteria as query sequences revealed that R. l. bv. viciae 3841 has ten putative protein secretion systems. These are the general export pathway (GEP), a twin-arginine translocase (TAT) secretion system, four separate Type I systems, one putative Type IV system and three Type V autotransporters. Mutations in genes encoding each of these (except the GEP) were generated, but only mutations affecting the PrsDE (Type I) and TAT systems were observed to affect the growth phenotype and the profile of proteins in the culture supernatant. Bioinformatic analysis and mass fingerprinting of tryptic fragments of culture supernatant proteins identified 14 putative Type I substrates, 12 of which are secreted via the PrsDE, secretion system. The TAT mutant was defective for the symbiosis, forming nodules incapable of nitrogen fixation.
Conclusion: None of the R. l. bv. viciae 3841 protein secretion systems putatively involved in the secretion of proteins to the extracellular space (Type I, Type IV, Type V) is required for establishing the symbiosis with legumes. The PrsDE (Type I) system was shown to be the major route of protein secretion in non-symbiotic cells and to secrete proteins of widely varied size and predicted function. This is in contrast to many Type I systems from other bacteria, which typically secrete specific substrates encoded by genes often localised in close proximity to the genes encoding the secretion system itself.
Figures



References
-
- Downie JA, Surin BP. Either of two nod gene loci can complement the nodulation defect of a nod deletion mutant of Rhizobium leguminosarum bv. viciae. Mol Gen Genet. 1990;222:81–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

