Evidence of different metabolic phenotypes in humans
- PMID: 18230739
- PMCID: PMC2234159
- DOI: 10.1073/pnas.0705685105
Evidence of different metabolic phenotypes in humans
Abstract
The study of metabolic responses to drugs, environmental changes, and diseases is a new promising area of metabonomic research. Metabolic fingerprints can be obtained by analytical techniques such as nuclear magnetic resonance (NMR). In principle, alterations of these fingerprints due to appearance/disappearance or concentration changes of metabolites can provide early evidences of, for example, onset of diseases. A major drawback in this approach is the strong day-to-day variability of the individual metabolic fingerprint, which should be rather called a metabolic "snapshot." We show here that a thorough statistical analysis performed on NMR spectra of human urine samples reveals an invariant part characteristic of each person, which can be extracted from the analysis of multiple samples of each single subject. This finding (i) provides evidence that individual metabolic phenotypes may exist and (ii) opens new perspectives to metabonomic studies, based on the possibility of eliminating the daily "noise" by multiple sample collection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
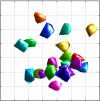
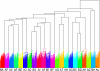
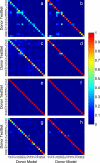

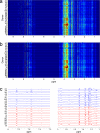
References
-
- Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. - PubMed
-
- Gavaghan CL, Holmes E, Lenz E, Wilson ID, Nicholson JK. FEBS Lett. 2000;484:169–174. - PubMed
-
- Rezzi S, Ramadan Z, Fay LB, Kochhar S. J Proteome Res. 2007;6:513–525. - PubMed
-
- Kussmann M, Raymond F, Affolter M. J Biotechnol. 2006;124:758–787. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

