The RNA binding protein Hfq interacts specifically with tRNAs
- PMID: 18230766
- PMCID: PMC2248270
- DOI: 10.1261/rna.531408
The RNA binding protein Hfq interacts specifically with tRNAs
Abstract
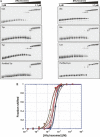
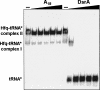
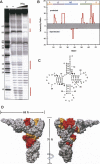
Hfq is an RNA binding protein that has been studied extensively for its role in the biology of small noncoding RNAs (ncRNAs) in bacteria, where it facilitates post-transcriptional gene regulation during stress responses. We show that Hfq also binds with high specificity and nanomolar affinity to tRNAs despite their lack of a canonical A/U rich single-stranded sequence. This affinity is comparable to that of Hfq for its validated ncRNA targets. Two sites on tRNAs are protected by Hfq binding, one on the D-stem and the other on the T-stem. Mutational analysis and competitive binding experiments indicate that Hfq uses its proximal surface (also called the L4 face) to bind tRNAs, the same surface that interacts with ncRNAs but a site distinct from where poly(A) oligonucleotides bind. hfq knockout strains are known to have broad pleiotropic phenotypes, but none of them are easily explained by or imply a role for tRNA binding. We show that hfq deletion strains have a previously unrecognized phenotype associated with mistranslation and significantly reduced translational fidelity. We infer that tRNA binding and reduced fidelity are linked by a role for Hfq in tRNA modification.
Figures
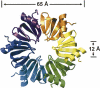
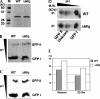
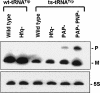
Similar articles
-
Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites.Biochemistry. 2006 Apr 18;45(15):4875-87. doi: 10.1021/bi0523613. Biochemistry. 2006. PMID: 16605255
-
Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS.RNA. 2013 Aug;19(8):1089-104. doi: 10.1261/rna.034595.112. Epub 2013 Jun 26. RNA. 2013. PMID: 23804244 Free PMC article.
-
C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation.J Mol Biol. 2010 Nov 26;404(2):173-82. doi: 10.1016/j.jmb.2010.09.038. Epub 2010 Oct 1. J Mol Biol. 2010. PMID: 20888338
-
New molecular interactions broaden the functions of the RNA chaperone Hfq.Curr Genet. 2019 Dec;65(6):1313-1319. doi: 10.1007/s00294-019-00990-y. Epub 2019 May 18. Curr Genet. 2019. PMID: 31104083 Review.
-
Cycling of RNAs on Hfq.RNA Biol. 2013 Apr;10(4):619-26. doi: 10.4161/rna.24044. Epub 2013 Mar 6. RNA Biol. 2013. PMID: 23466677 Free PMC article. Review.
Cited by
-
Conserved arginines on the rim of Hfq catalyze base pair formation and exchange.Nucleic Acids Res. 2013 Aug;41(15):7536-46. doi: 10.1093/nar/gkt521. Epub 2013 Jun 14. Nucleic Acids Res. 2013. PMID: 23771143 Free PMC article.
-
Polyadenylation helps regulate functional tRNA levels in Escherichia coli.Nucleic Acids Res. 2012 May;40(10):4589-603. doi: 10.1093/nar/gks006. Epub 2012 Jan 28. Nucleic Acids Res. 2012. PMID: 22287637 Free PMC article.
-
Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts.Nucleic Acids Res. 2010 Jun;38(11):3794-808. doi: 10.1093/nar/gkq032. Epub 2010 Mar 26. Nucleic Acids Res. 2010. PMID: 20348540 Free PMC article.
-
Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E.EMBO J. 2017 Feb 1;36(3):374-387. doi: 10.15252/embj.201694639. Epub 2016 Nov 11. EMBO J. 2017. PMID: 27836995 Free PMC article.
-
tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization.Cell Mol Life Sci. 2010 May;67(9):1447-63. doi: 10.1007/s00018-010-0271-4. Epub 2010 Feb 14. Cell Mol Life Sci. 2010. PMID: 20155482 Free PMC article. Review.
References
-
- Arluison, V., Derreumaux, P., Allemand, F., Folichon, M., Hajnsdorf, E., Regnier, P. Structural modeling of the Sm-like protein Hfq from Escherichia coli . J. Mol. Biol. 2002;320:705–712. - PubMed
-
- Bjork, G.R. Stable RNA modification. In Escherichia coli and Salmonella: Cellular and molecular biology . In: Neidhardt F.C., et al., editors. 2d ed. Vol. 1. ASM Press; Washington, DC: 1996. pp. 861–886.
-
- Butland, G., Peregrin-Alvarez, J.M., Li, J., Yang, W., Yang, X., Canadien, V., Starostine, A., Richards, D., Beattie, B., Krogan, N., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli . Nature. 2005;433:531–537. - PubMed
-
- Cabello-Villegas, J., Winkler, M.E., Nikonowicz, E.P. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem–loops of Escherichia coli tRNAPhe . J. Mol. Biol. 2002;319:1015–1034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
