Evolution of protein domain promiscuity in eukaryotes
- PMID: 18230802
- PMCID: PMC2259109
- DOI: 10.1101/gr.6943508
Evolution of protein domain promiscuity in eukaryotes
Abstract
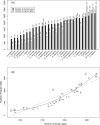
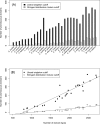
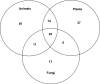
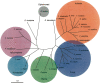
Numerous eukaryotic proteins contain multiple domains. Certain domains show a tendency to occur in diverse domain architectures and can be considered "promiscuous." These promiscuous domains are, typically, involved in protein-protein interactions and play crucial roles in interaction networks, particularly those that contribute to signal transduction. A systematic comparative-genomic analysis of promiscuous domains in eukaryotes is described. Two quantitative measures of domain promiscuity are introduced and applied to the analysis of 28 genomes of diverse eukaryotes. Altogether, 215 domains are identified as strongly promiscuous. The fraction of promiscuous domains in animals is shown to be significantly greater than that in fungi or plants. Evolutionary reconstructions indicate that domain promiscuity is a volatile, relatively fast-changing feature of eukaryotic proteins, with few domains remaining promiscuous throughout the evolution of eukaryotes. Some domains appear to have attained promiscuity independently in different lineages, for example, animals and plants. It is proposed that promiscuous domains persist within a relatively small pool of evolutionarily stable domain combinations from which numerous rare architectures emerge during evolution. Domain promiscuity positively correlates with the number of experimentally detected domain interactions and with the strength of purifying selection affecting a domain. Thus, evolution of promiscuous domains seems to be constrained by the diversity of their interaction partners. The set of promiscuous domains is enriched for domains mediating protein-protein interactions that are involved in various forms of signal transduction, especially in the ubiquitin system and in chromatin. Thus, a limited repertoire of promiscuous domains makes a major contribution to the diversity and evolvability of eukaryotic proteomes and signaling networks.
Figures

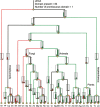
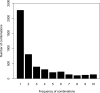

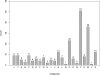
References
-
- Anantharaman V., Koonin E.V., Aravind L., Koonin E.V., Aravind L., Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. - PubMed
-
- Apic G., Gough J., Teichmann S.A., Gough J., Teichmann S.A., Teichmann S.A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001;310:311–325. - PubMed
-
- Aravind L., Dixit V.M., Koonin E.V., Dixit V.M., Koonin E.V., Koonin E.V. Apoptotic molecular machinery: Vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. - PubMed
-
- Bashton M., Chothia C., Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. - PubMed
-
- Bohning D., Schlattmann P., Lindsay B., Schlattmann P., Lindsay B., Lindsay B. Computer-assisted analysis of mixtures (C.A.MAM): Statistical algorithms. Biometrics. 1992;48:283–303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
