A transcriptional fingerprint of estrogen in human breast cancer predicts patient survival
- PMID: 18231641
- PMCID: PMC2213902
- DOI: 10.1593/neo.07859
A transcriptional fingerprint of estrogen in human breast cancer predicts patient survival
Abstract
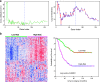
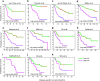
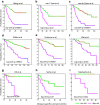
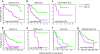
Estrogen signaling plays an essential role in breast cancer progression, and estrogen receptor (ER) status has long been a marker of hormone responsiveness. However, ER status alone has been an incomplete predictor of endocrine therapy, as some ER+ tumors, nevertheless, have poor prognosis. Here we sought to use expression profiling of ER+ breast cancer cells to screen for a robust estrogen-regulated gene signature that may serve as a better indicator of cancer outcome. We identified 532 estrogen-induced genes and further developed a 73-gene signature that best separated a training set of 286 primary breast carcinomas into prognostic subtypes by stepwise cross-validation. Notably, this signature predicts clinical outcome in over 10 patient cohorts as well as their respective ER+ subcohorts. Further, this signature separates patients who have received endocrine therapy into two prognostic subgroups, suggesting its specificity as a measure of estrogen signaling, and thus hormone sensitivity. The 73-gene signature also provides additional predictive value for patient survival, independent of other clinical parameters, and outperforms other previously reported molecular outcome signatures. Taken together, these data demonstrate the power of using cell culture systems to screen for robust gene signatures of clinical relevance.
Figures

References
-
- Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344:276–285. - PubMed
-
- Jordan VC. Tamoxifen: a most unlikely pioneering medicine. Nat Rev Drug Discov. 2003;2:205–213. - PubMed
-
- Early Breast Cancer Trialists' Collaborative Group (EBCTCG), author Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. - PubMed
-
- Brenner H. Long-term survival rates of cancer patients achieved by the end of the 20th century: a period analysis. Lancet. 2002;360:1131–1135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
