Strong purifying selection in transmission of mammalian mitochondrial DNA
- PMID: 18232733
- PMCID: PMC2214808
- DOI: 10.1371/journal.pbio.0060010
Strong purifying selection in transmission of mammalian mitochondrial DNA
Abstract
There is an intense debate concerning whether selection or demographics has been most important in shaping the sequence variation observed in modern human mitochondrial DNA (mtDNA). Purifying selection is thought to be important in shaping mtDNA sequence evolution, but the strength of this selection has been debated, mainly due to the threshold effect of pathogenic mtDNA mutations and an observed excess of new mtDNA mutations in human population data. We experimentally addressed this issue by studying the maternal transmission of random mtDNA mutations in mtDNA mutator mice expressing a proofreading-deficient mitochondrial DNA polymerase. We report a rapid and strong elimination of nonsynonymous changes in protein-coding genes; the hallmark of purifying selection. There are striking similarities between the mutational patterns in our experimental mouse system and human mtDNA polymorphisms. These data show strong purifying selection against mutations within mtDNA protein-coding genes. To our knowledge, our study presents the first direct experimental observations of the fate of random mtDNA mutations in the mammalian germ line and demonstrates the importance of purifying selection in shaping mitochondrial sequence diversity.
Conflict of interest statement
Figures
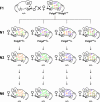
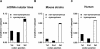
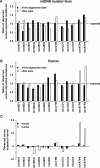
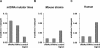
References
-
- Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. - PubMed
-
- Shoubridge EA, Wai T. Mitochondrial DNA and the mammalian oocyte. Curr Top Dev Biol. 2007;77:87–111. - PubMed
-
- Enriquez JA, Cabezas-Herrera J, Bayona-Bafaluy MP, Attardi G. Very rare complementation between mitochondria carrying different mitochondrial DNA mutations points to intrinsic genetic autonomy of the organelles in cultured human cells. J Biol Chem. 2000;275:11207–11215. - PubMed
-
- Piganeau G, Eyre-Walker A. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity. 2004;92:282–288. - PubMed
-
- Elson JL, Lightowlers RN. Mitochondrial DNA clonality in the dock: can surveillance swing the case? Trends Genet. 2006;22:603–607. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

