PEPOP: computational design of immunogenic peptides
- PMID: 18234071
- PMCID: PMC2262870
- DOI: 10.1186/1471-2105-9-71
PEPOP: computational design of immunogenic peptides
Abstract
Background: Most methods available to predict protein epitopes are sequence based. There is a need for methods using 3D information for prediction of discontinuous epitopes and derived immunogenic peptides.
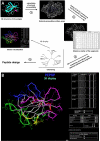
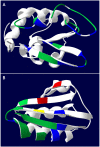
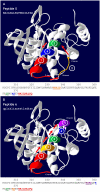
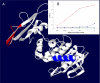
Results: PEPOP uses the 3D coordinates of a protein both to predict clusters of surface accessible segments that might correspond to epitopes and to design peptides to be used to raise antibodies that target the cognate antigen at specific sites. To verify the ability of PEPOP to identify epitopes, 13 crystallographically defined epitopes were compared with PEPOP clusters: specificity ranged from 0.75 to 1.00, sensitivity from 0.33 to 1.00, and the positive predictive value from 0.19 to 0.89. Comparison of these results with those obtained with two other prediction algorithms showed comparable specificity and slightly better sensitivity and PPV. To prove the capacity of PEPOP to predict immunogenic peptides that induce protein cross-reactive antibodies, several peptides were designed from the 3D structure of model antigens (IA-2, TPO, and IL8) and chemically synthesized. The reactivity of the resulting anti-peptides antibodies with the cognate antigens was measured. In 80% of the cases (four out of five peptides), the flanking protein sequence process (sequence-based) of PEPOP successfully proposed peptides that elicited antibodies cross-reacting with the parent proteins. Polyclonal antibodies raised against peptides designed from amino acids which are spatially close in the protein, but separated in the sequence, could also be obtained, although they were much less reactive. The capacity of PEPOP to design immunogenic peptides that induce antibodies suitable for a sandwich capture assay was also demonstrated.
Conclusion: PEPOP has the potential to guide experimentalists that want to localize an epitope or design immunogenic peptides for raising antibodies which target proteins at specific sites. More successful predictions of immunogenic peptides were obtained when a peptide was continuous as compared with peptides corresponding to discontinuous epitopes. PEPOP is available for use at http://diagtools.sysdiag.cnrs.fr/PEPOP/.
Figures
References
-
- Reineke U, Kramer A, Schneider-Mergener J. Antigen sequence- and library-based mapping of linear and discontinuous protein-protein-interaction sites by spot synthesis. Curr Top Microbiol Immunol. 1999;243:23–36. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

