Analysis of non-TIR NBS-LRR resistance gene analogs in Musa acuminata Colla: isolation, RFLP marker development, and physical mapping
- PMID: 18234103
- PMCID: PMC2262081
- DOI: 10.1186/1471-2229-8-15
Analysis of non-TIR NBS-LRR resistance gene analogs in Musa acuminata Colla: isolation, RFLP marker development, and physical mapping
Abstract
Background: Many commercial banana varieties lack sources of resistance to pests and diseases, as a consequence of sterility and narrow genetic background. Fertile wild relatives, by contrast, possess greater variability and represent potential sources of disease resistance genes (R-genes). The largest known family of plant R-genes encode proteins with nucleotide-binding site (NBS) and C-terminal leucine-rich repeat (LRR) domains. Conserved motifs in such genes in diverse plant species offer a means for isolation of candidate genes in banana which may be involved in plant defence.
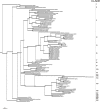
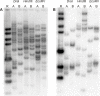
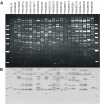
Results: A computational strategy was developed for unbiased conserved motif discovery in NBS and LRR domains in R-genes and homologues in monocotyledonous plant species. Degenerate PCR primers targeting conserved motifs were tested on the wild cultivar Musa acuminata subsp. burmannicoides, var. Calcutta 4, which is resistant to a number of fungal pathogens and nematodes. One hundred and seventy four resistance gene analogs (RGAs) were amplified and assembled into 52 contiguous sequences. Motifs present were typical of the non-TIR NBS-LRR RGA subfamily. A phylogenetic analysis of deduced amino-acid sequences for 33 RGAs with contiguous open reading frames (ORFs), together with RGAs from Arabidopsis thaliana and Oryza sativa, grouped most Musa RGAs within monocotyledon-specific clades. RFLP-RGA markers were developed, with 12 displaying distinct polymorphisms in parentals and F1 progeny of a diploid M. acuminata mapping population. Eighty eight BAC clones were identified in M. acuminata Calcutta 4, M. acuminata Grande Naine, and M. balbisiana Pisang Klutuk Wulung BAC libraries when hybridized to two RGA probes. Multiple copy RGAs were common within BAC clones, potentially representing variation reservoirs for evolution of new R-gene specificities.
Conclusion: This is the first large scale analysis of NBS-LRR RGAs in M. acuminata Calcutta 4. Contig sequences were deposited in GenBank and assigned numbers ER935972 - ER936023. RGA sequences and isolated BACs are a valuable resource for R-gene discovery, and in future applications will provide insight into the organization and evolution of NBS-LRR R-genes in the Musa A and B genome. The developed RFLP-RGA markers are applicable for genetic map development and marker assisted selection for defined traits such as pest and disease resistance.
Figures



Similar articles
-
Isolation, genetic variation and expression of TIR-NBS-LRR resistance gene analogs from western white pine ( Pinus monticola Dougl. ex. D. Don.).Mol Genet Genomics. 2003 Dec;270(5):432-41. doi: 10.1007/s00438-003-0940-1. Epub 2003 Oct 28. Mol Genet Genomics. 2003. PMID: 14586641
-
Genomes, diversity and resistance gene analogues in Musa species.Cytogenet Genome Res. 2008;121(1):59-66. doi: 10.1159/000124383. Epub 2008 May 7. Cytogenet Genome Res. 2008. PMID: 18544928
-
Isolation of TIR and non-TIR NBS--LRR resistance gene analogues and identification of molecular markers linked to a powdery mildew resistance locus in chestnut rose (Rosa roxburghii Tratt).Theor Appl Genet. 2005 Sep;111(5):819-30. doi: 10.1007/s00122-005-0002-7. Epub 2005 Oct 18. Theor Appl Genet. 2005. PMID: 16075209
-
Disease Resistance Gene Analogs (RGAs) in Plants.Int J Mol Sci. 2015 Aug 14;16(8):19248-90. doi: 10.3390/ijms160819248. Int J Mol Sci. 2015. PMID: 26287177 Free PMC article. Review.
-
Identification and expression profiling analysis of NBS-LRR genes involved in Fusarium oxysporum f.sp. conglutinans resistance in cabbage.3 Biotech. 2019 May;9(5):202. doi: 10.1007/s13205-019-1714-8. Epub 2019 May 4. 3 Biotech. 2019. PMID: 31065502 Free PMC article. Review.
Cited by
-
Identification of Differentially-Expressed Genes in Response to Mycosphaerella fijiensis in the Resistant Musa Accession 'Calcutta-4' Using Suppression Subtractive Hybridization.PLoS One. 2016 Aug 3;11(8):e0160083. doi: 10.1371/journal.pone.0160083. eCollection 2016. PLoS One. 2016. PMID: 27487237 Free PMC article.
-
Mechanisms of haplotype divergence at the RGA08 nucleotide-binding leucine-rich repeat gene locus in wild banana (Musa balbisiana).BMC Plant Biol. 2010 Jul 16;10:149. doi: 10.1186/1471-2229-10-149. BMC Plant Biol. 2010. PMID: 20637079 Free PMC article.
-
Metabolite profiling characterises chemotypes of Musa diploids and triploids at juvenile and pre-flowering growth stages.Sci Rep. 2019 Mar 15;9(1):4657. doi: 10.1038/s41598-019-41037-z. Sci Rep. 2019. PMID: 30874619 Free PMC article.
-
Evolution and Conservation of Plant NLR Functions.Front Immunol. 2013 Sep 25;4:297. doi: 10.3389/fimmu.2013.00297. Front Immunol. 2013. PMID: 24093022 Free PMC article. Review.
-
Development of expressed sequence tag and expressed sequence tag-simple sequence repeat marker resources for Musa acuminata.AoB Plants. 2012;2012:pls030. doi: 10.1093/aobpla/pls030. Epub 2012 Nov 26. AoB Plants. 2012. PMID: 23240072 Free PMC article.
References
-
- Frison EA, Sharrock S. In: The Economic, social and nutritional importance of banana in the world: 1998/11/10. C.Picq , E.Fouré and E.A.Frison. , editor. Douala, Cameroon, International Network for the Improvement of Banana and Plantain; 1999. pp. 21–31. (Proceedings of the Bananas and Food Security).
-
- Janick J. Acta Hort (ISHS) Vol. 490. Tenerife, Spain, International Society for Horticultural Science, Leuven (BEL); 1998. Fruit breeding in the 21st century: 1997/11/10; Tenerife (ESP). pp. 39–46. (First International Symposium on Banana in the Subtropics).
-
- HH F. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. doi: 10.1146/annurev.py.09.090171.001423. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

