Structural Insights into ribosome recycling factor interactions with the 70S ribosome
- PMID: 18234219
- PMCID: PMC2712656
- DOI: 10.1016/j.jmb.2007.12.048
Structural Insights into ribosome recycling factor interactions with the 70S ribosome
Abstract
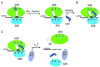
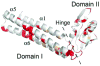
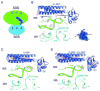
At the end of translation in bacteria, ribosome recycling factor (RRF) is used together with elongation factor G to recycle the 30S and 50S ribosomal subunits for the next round of translation. In x-ray crystal structures of RRF with the Escherichia coli 70S ribosome, RRF binds to the large ribosomal subunit in the cleft that contains the peptidyl transferase center. Upon binding of either E. coli or Thermus thermophilus RRF to the E. coli ribosome, the tip of ribosomal RNA helix 69 in the large subunit moves away from the small subunit toward RRF by 8 A, thereby disrupting a key contact between the small and large ribosomal subunits termed bridge B2a. In the ribosome crystals, the ability of RRF to destabilize bridge B2a is influenced by crystal packing forces. Movement of helix 69 involves an ordered-to-disordered transition upon binding of RRF to the ribosome. The disruption of bridge B2a upon RRF binding to the ribosome seen in the present structures reveals one of the key roles that RRF plays in ribosome recycling, the dissociation of 70S ribosomes into subunits. The structures also reveal contacts between domain II of RRF and protein S12 in the 30S subunit that may also play a role in ribosome recycling.
Figures





References
-
- Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A. The ribosome-recycling step: consensus or controversy? Trends Biochem Sci. 2006;31:143–9. - PubMed
-
- Hirashima A, Kaji A. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J Biol Chem. 1973;248:7580–7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

