Nbs1-dependent binding of Mre11 to adenovirus E4 mutant viral DNA is important for inhibiting DNA replication
- PMID: 18234271
- PMCID: PMC2494955
- DOI: 10.1016/j.virol.2007.12.034
Nbs1-dependent binding of Mre11 to adenovirus E4 mutant viral DNA is important for inhibiting DNA replication
Abstract
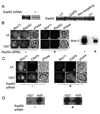
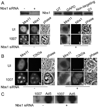
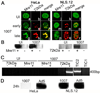
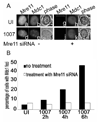
Adenovirus (Ad) infections stimulate the activation of cellular DNA damage response and repair pathways. Ad early regulatory proteins prevent activation of DNA damage responses by targeting the MRN complex, composed of the Mre11, Rad50 and Nbs1 proteins, for relocalization and degradation. In the absence of these viral proteins, Mre11 colocalizes with viral DNA replication foci. Mre11 foci formation at DNA damage induced by ionizing radiation depends on the Nbs1 component of the MRN complex and is stabilized by the mediator of DNA damage checkpoint protein 1 (Mdc1). We find that Nbs1 is required for Mre11 localization at DNA replication foci in Ad E4 mutant infections. Mre11 is important for Mdc1 foci formation in infected cells, consistent with its role as a sensor of DNA damage. Chromatin immunoprecipitation assays indicate that both Mre11 and Mdc1 are physically bound to viral DNA, which could account for their localization in viral DNA containing foci. Efficient binding of Mre11 to E4 mutant DNA depends on the presence of Nbs1, and is correlated with a significant E4 mutant DNA replication defect. Our results are consistent with a model in which physical interaction of Mre11 with viral DNA is mediated by Nbs1, and interferes with viral DNA replication.
Figures

References
-
- Aspegren A, Rabino C, Bridge E. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 1998;245:203–213. - PubMed
-
- Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;63:631–638. - PubMed
-
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

