Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions
- PMID: 18234722
- PMCID: PMC2655627
- DOI: 10.1242/dev.014316
Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions
Abstract
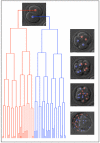
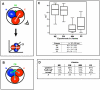
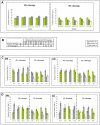
Setting aside pluripotent cells that give rise to the future body is a central cell fate decision in mammalian development. It requires that some blastomeres divide asymmetrically to direct cells to the inside of the embryo. Despite its importance, it is unknown whether the decision to divide symmetrically versus asymmetrically shows any spatial or temporal pattern, whether it is lineage-dependent or occurs at random, or whether it influences the orientation of the embryonic-abembryonic axis. To address these questions, we developed time-lapse microscopy to enable a complete 3D analysis of the origins, fates and divisions of all cells from the 2- to 32-cell blastocyst stage. This showed how in the majority of embryos, individual blastomeres give rise to distinct blastocyst regions. Tracking the division orientation of all cells revealed a spatial and temporal relationship between symmetric and asymmetric divisions and how this contributes to the generation of inside and outside cells and thus embryo patterning. We found that the blastocyst cavity, defining the abembryonic pole, forms where symmetric divisions predominate. Tracking cell ancestry indicated that the pattern of symmetric/asymmetric divisions of a blastomere can be influenced by its origin in relation to the animal-vegetal axis of the zygote. Thus, it appears that the orientation of the embryonic-abembryonic axis is anticipated by earlier cell division patterns. Together, our results suggest that two steps influence the allocation of cells to the blastocyst. The first step, involving orientation of 2- to 4-cell divisions along the animal-vegetal axis, can affect the second step, the establishment of inside and outside cell populations by asymmetric 8- to 32-cell divisions.
Figures


References
-
- Alarcon VB, Marikawa Y. Deviation of the blastocyst axis from the first cleavage plane does not affect the quality of mouse postimplantation development. Biol Reprod. 2003;69:1208–12. - PubMed
-
- Barlow P, Owen DAJ, Graham CF. DNA synthesis in the preimplantation mouse embryo. J. Embryol. Exp. Morphol. 1972;27:431–435. - PubMed
-
- Chroscicka A, Komorowski S, Maleszewski M. Both blastomeres of the mouse 2-cell embryo contribute to the embryonic portion of the blastocyst. Mol Reprod Dev. 2004;68:308–12. - PubMed
-
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–31. - PubMed
-
- Fleming TP. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Dev Biol. 1987;119:520–31. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

