Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis
- PMID: 18234864
- PMCID: PMC2292969
- DOI: 10.1128/JCM.02089-07
Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis
Abstract
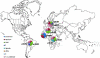
Standardized mycobacterial interspersed repetitive-unit-variable-number tandem repeat (MIRU-VNTR) typing based on 15 and 24 loci recently has been proposed for Mycobacterium tuberculosis genotyping. So far, this optimized system has been assessed in a single, 1-year population-based study performed in Germany (M. C. Oelemann, R. Diel, V. Vatin, W. Haas, S. Rusch-Gerdes, C. Locht, S. Niemann, and P. Supply, J. Clin. Microbiol. 45:691-697, 2007). Here, we evaluated these optimized formats in a much larger population-based study conducted during 39 months in the Brussels capital region of Belgium. Isolates from 807 patients were genotyped. The resolution power, cluster, and lineage identification by the standardized MIRU-VNTR sets were compared to those obtained using standardized IS6110-restriction fragment length polymorphism (RFLP), spoligotyping, and a previous 12-MIRU-VNTR-locus set. On a subset representing 77% of the cases during a 16-month period, a high concordance was observed between unique isolates or strain clusters as defined by standardized MIRU-VNTR and IS6110-RFLP (i.e., more than five IS6110 bands). When extended to the entire population-based collection, the discriminatory subset of 15 loci decreased the strain-clustering rate by almost twofold compared to that of the old 12-locus set. The addition of the nine ancillary MIRU-VNTR loci and/or spoligotyping only slightly further decreased this strain-clustering rate. Familial, social, and/or geographic proximity links were found in 48% of the clusters identified, and well-known risk factors for tuberculosis transmission were identified. Finally, an excellent correspondence was determined between our MIRU-VNTR-spoligotyping strain identifications and external reference strain lineages included in the MIRU-VNTRplus database and identified by, e.g., large sequence polymorphisms. Our results reinforce the proposal of standardized MIRU-VNTR typing as a new reference genotyping method for the epidemiological and phylogenetic screening of M. tuberculosis strains.
Figures

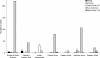
References
-
- Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus Yersinia pestis. Proc. Natl. Acad. Sci. USA 10117837-17842. - PMC - PubMed
-
- Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 3301710-1716. - PubMed
-
- Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39783-789. - PubMed
-
- Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 3491149-1156. - PubMed
-
- Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 1045-52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

