Characterization of metal binding in the active sites of acireductone dioxygenase isoforms from Klebsiella ATCC 8724
- PMID: 18237192
- PMCID: PMC2267756
- DOI: 10.1021/bi7004152
Characterization of metal binding in the active sites of acireductone dioxygenase isoforms from Klebsiella ATCC 8724
Abstract
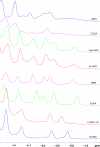
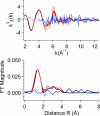
The two acireductone dioxygenase (ARD) isozymes from the methionine salvage pathway of Klebsiella ATCC 8724 present an unusual case in which two enzymes with different structures and distinct activities toward their common substrates (1,2-dihydroxy-3-oxo-5-(methylthio)pent-1-ene and dioxygen) are derived from the same polypeptide chain. Structural and functional differences between the two isozymes are determined by the type of M2+ metal ion bound in the active site. The Ni2+-bound NiARD catalyzes an off-pathway shunt from the methionine salvage pathway leading to the production of formate, methylthiopropionate, and carbon monoxide, while the Fe2+-bound FeARD' catalyzes the on-pathway formation of methionine precursor 2-keto-4-methylthiobutyrate and formate. Four potential protein-based metal ligands were identified by sequence homology and structural considerations. Based on the results of site-directed mutagenesis experiments, X-ray absorption spectroscopy (XAS), and isothermal calorimetry measurements, it is concluded that the same four residues, His96, His98, Glu102 and His140, provide the protein-based ligands for the metal in both the Ni- and Fe-containing forms of the enzyme, and subtle differences in the local backbone conformations trigger the observed structural and functional differences between the FeARD' and NiARD isozymes. Furthermore, both forms of the enzyme bind their respective metals with pseudo-octahedral geometry, and both may lose a histidine ligand upon binding of substrate under anaerobic conditions. However, mutations at two conserved nonligand acidic residues, Glu95 and Glu100, result in low metal contents for the mutant proteins as isolated, suggesting that some of the conserved charged residues may aid in transfer of metal from in vivo sources or prevent the loss of metal to stronger chelators. The Glu100 mutant reconstitutes readily but has low activity. Mutation of Asp101 results in an active enzyme that incorporates metal in vivo but shows evidence of mixed forms.
Figures





References
-
- Schlenk F. Methylthioadenosine. Adv. Enzymol. 1983;54:195–265. - PubMed
-
- Oredsson SM. Polyamine dependence of normal cell-cycle progression. Biochem. Soc. Trans. 2003;31:366–370. - PubMed
-
- Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Ann. Rev. Pharmacol. Toxicol. 1995;35:55–91. - PubMed
-
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy. Cancer Res. 1988;48:759–774. - PubMed
-
- Shapiro SK, Barrett A. 5-Methylthioribose as a precursor of the carbon chain of methionine. Biochem. Biophys. Res. Comm. 1981;102:302–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

