Identification of a novel Leucine-rich repeat protein and candidate PP1 regulatory subunit expressed in developing spermatids
- PMID: 18237440
- PMCID: PMC2270827
- DOI: 10.1186/1471-2121-9-9
Identification of a novel Leucine-rich repeat protein and candidate PP1 regulatory subunit expressed in developing spermatids
Abstract
Background: Spermatogenesis is comprised of a series of highly regulated developmental changes that transform the precursor germ cell into a highly specialized spermatozoon. The last phase of spermatogenesis, termed spermiogenesis, involves dramatic morphological change including formation of the acrosome, elongation and condensation of the nucleus, formation of the flagella, and disposal of unnecessary cytoplasm. A prominent cytoskeletal component of the developing spermatid is the manchette, a unique microtubular structure that surrounds the nucleus of the developing spermatid and is thought to assist in both the reshaping of the nucleus and redistribution of spermatid cytoplasm. Although the molecular motor KIFC1 has been shown to associate with the manchette, its precise role in function of the manchette and the identity of its testis specific protein partners are unknown. The purpose of this study was to identify proteins in the testis that interact with KIFC1 using a yeast 2 hybrid screen of a testis cDNA library.
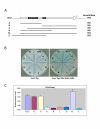
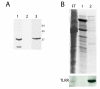
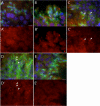
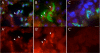
Results: Thirty percent of the interacting clones identified in our screen contain an identical cDNA encoding a 40 kD protein. This interacting protein has 4 leucine-rich repeats in its amino terminal half and is expressed primarily in the testis; therefore we have named this protein testis leucine-rich repeat protein or TLRR. TLRR was also found to associate tightly with the KIFC1 targeting domain using affinity chromatography. In addition to the leucine-rich repeats, TLRR contains a consensus-binding site for protein phosphatase-1 (PP1). Immunocytochemistry using a TLRR specific antibody demonstrates that this protein is found near the manchette of developing spermatids.
Conclusion: We have identified a previously uncharacterized leucine-rich repeat protein that is expressed abundantly in the testis and associates with the manchette of developing spermatids, possibly through its interaction with the KIFC1 molecular motor. TLRR is homologous to a class of regulatory subunits for PP1, a central phosphatase in the reversible phosphorylation of proteins that is key to modulation of many intracellular processes. TLRR may serve to target this important signaling molecule near the nucleus of developing spermatids in order to control the cellular rearrangements of spermiogenesis.
Figures


References
-
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52:198–236. - PubMed
-
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

