Structure and specificity of lamprey monoclonal antibodies
- PMID: 18238899
- PMCID: PMC2542867
- DOI: 10.1073/pnas.0711619105
Structure and specificity of lamprey monoclonal antibodies
Abstract
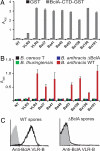
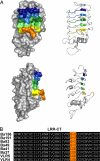
Adaptive immunity in jawless vertebrates (lamprey and hagfish) is mediated by lymphocytes that undergo combinatorial assembly of leucine-rich repeat (LRR) gene segments to create a diverse repertoire of variable lymphocyte receptor (VLR) genes. Immunization with particulate antigens induces VLR-B-bearing lymphocytes to secrete antigen-specific VLR-B antibodies. Here, we describe the production of recombinant VLR-B antibodies specific for BclA, a major coat protein of Bacillus anthracis spores. The recombinant VLR-B antibodies possess 8-10 uniform subunits that collectively bind antigen with high avidity. Sequence analysis, mutagenesis, and modeling studies show that antigen binding involves residues in the beta-sheets lining the VLR-B concave surface. EM visualization reveals tetrameric and pentameric molecules having a central core and highly flexible pairs of stalk-region "arms" with antigen-binding "hands." Remarkable antigen-binding specificity, avidity, and stability predict that these unusual LRR-based monoclonal antibodies will find many biomedical uses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. - PubMed
-
- Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–656. - PubMed
-
- Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. - PubMed
-
- Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8:206–213. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

