Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate
- PMID: 18239134
- PMCID: PMC2254926
- DOI: 10.1105/tpc.107.052001
Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate
Abstract
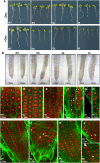
The Ypt3/Rab11/Rab25 subfamily of Rab GTPases has expanded greatly in Arabidopsis thaliana, comprising 26 members in six provisional subclasses, Rab-A1 to Rab-A6. We show that the Rab-A2 and Rab-A3 subclasses define a novel post-Golgi membrane domain in Arabidopsis root tips. The Rab-A2/A3 compartment was distinct from but often close to Golgi stacks and prevacuolar compartments and partly overlapped the VHA-a1 trans-Golgi compartment. It was also sensitive to brefeldin A and accumulated FM4-64 before prevacuolar compartments did. Mutations in RAB-A2a that were predicted to stabilize the GDP- or GTP-bound state shifted the location of the protein to the Golgi or plasma membrane, respectively. In mitosis, KNOLLE accumulated principally in the Rab-A2/A3 compartment. During cytokinesis, Rab-A2 and Rab-A3 proteins localized precisely to the growing margins of the cell plate, but VHA-a1, GNOM, and prevacuolar markers were excluded. Inducible expression of dominant-inhibitory mutants of RAB-A2a resulted in enlarged, polynucleate, meristematic cells with cell wall stubs. The Rab-A2/A3 compartment, therefore, is a trans-Golgi compartment that communicates with the plasma membrane and early endosomal system and contributes substantially to the cell plate. Despite the unique features of plant cytokinesis, membrane traffic to the division plane exhibits surprising molecular similarity across eukaryotic kingdoms in its reliance on Ypt3/Rab11/Rab-A GTPases.
Figures






References
-
- Albertson, R., Riggs, B., and Sullivan, W. (2005). Membrane traffic: A driving force in cytokinesis. Trends Cell Biol. 15 92–101. - PubMed
-
- Anai, T., Hasegawa, K., Watanabe, Y., Uchimiya, H., Ishizaki, R., and Matsui, M. (1991). Isolation and analysis of cDNAs encoding small GTP-binding proteins of Arabidopsis thaliana. Gene 108 259–264. - PubMed
-
- Ausubel, F., Brent, R., Kingston, R.E., Moore, J.G., Seidman, J.G., Smith, J.A., and Struhl, J.G. (1999). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
-
- Baluska, F., Liners, F., Hlavacka, A., Schlicht, M., Van Cutsem, P., McCurdy, D.W., and Menzel, D. (2005). Cell wall pectins and xyloglucans are internalized into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 225 141–155. - PubMed
-
- Baluska, F., Menzel, D., and Barlow, P.W. (2006). Cytokinesis in plant and animal cells: Endosomes ‘shut the door.’ Dev. Biol. 294 1–10. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

