The VSV polymerase can initiate at mRNA start sites located either up or downstream of a transcription termination signal but size of the intervening intergenic region affects efficiency of initiation
- PMID: 18241907
- PMCID: PMC2593140
- DOI: 10.1016/j.virol.2007.12.023
The VSV polymerase can initiate at mRNA start sites located either up or downstream of a transcription termination signal but size of the intervening intergenic region affects efficiency of initiation
Abstract
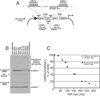
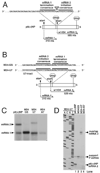
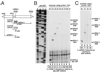
Transcription by the vesicular stomatitis virus (VSV) polymerase has been characterized as obligatorily sequential with transcription of each downstream gene dependent on termination of the gene immediately upstream. In studies described here we investigated the ability of the VSV RNA-dependent RNA polymerase (RdRp) to access mRNA initiation sites located at increasing distances either downstream or upstream of a transcription termination signal. Bi-cistronic subgenomic replicons were constructed containing progressively extended intergenic regions preceding the initiation site of a downstream gene. The ability of the RdRp to access the downstream sites was progressively reduced as the length of the intergenic region increased. Alternatively, bi-cistronic replicons were constructed containing an mRNA start signal located at increasing distances upstream of a termination site. Analysis of transcription of these "overlapped" genes showed that for an upstream mRNA start site to be recognized it had to contain not only the canonical 3'-UUGUCnnUAG-5' gene start signal, but that signal needed also to be preceded by a U7 tract. Access of these upstream mRNA initiation sites by the VSV RdRp was proportionately reduced with increasing distance between the termination site and the overlapped initiation signal. Possible mechanisms for how the RdRp accesses these upstream start sites are discussed.
Figures
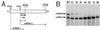

Similar articles
-
Identification of an upstream sequence element required for vesicular stomatitis virus mRNA transcription.J Virol. 2002 Aug;76(15):7632-41. doi: 10.1128/jvi.76.15.7632-7641.2002. J Virol. 2002. PMID: 12097577 Free PMC article.
-
Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts.J Virol. 1997 Mar;71(3):2127-37. doi: 10.1128/JVI.71.3.2127-2137.1997. J Virol. 1997. PMID: 9032346 Free PMC article.
-
cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation.J Virol. 1997 Nov;71(11):8718-25. doi: 10.1128/JVI.71.11.8718-8725.1997. J Virol. 1997. PMID: 9343230 Free PMC article.
-
Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus.Biochim Biophys Acta. 2002 Sep 13;1577(2):337-53. doi: 10.1016/s0167-4781(02)00462-1. Biochim Biophys Acta. 2002. PMID: 12213662 Review.
-
[The multifunctional RNA polymerase L protein of non-segmented negative strand RNA viruses catalyzes unique mRNA capping].Uirusu. 2014;64(2):165-78. doi: 10.2222/jsv.64.165. Uirusu. 2014. PMID: 26437839 Review. Japanese.
Cited by
-
Transcriptional Regulation in Ebola Virus: Effects of Gene Border Structure and Regulatory Elements on Gene Expression and Polymerase Scanning Behavior.J Virol. 2015 Dec 9;90(4):1898-909. doi: 10.1128/JVI.02341-15. Print 2016 Feb 15. J Virol. 2015. PMID: 26656691 Free PMC article.
-
A vesiculovirus showing a steepened transcription gradient and dominant trans-repression of virus transcription.J Virol. 2012 Aug;86(16):8884-9. doi: 10.1128/JVI.00358-12. Epub 2012 Jun 6. J Virol. 2012. PMID: 22674990 Free PMC article.
-
Selection for gene junction sequences important for VSV transcription.Virology. 2008 Oct 25;380(2):379-87. doi: 10.1016/j.virol.2008.08.001. Epub 2008 Sep 9. Virology. 2008. PMID: 18783810 Free PMC article.
-
Non-gradient and genotype-dependent patterns of RSV gene expression.PLoS One. 2020 Jan 10;15(1):e0227558. doi: 10.1371/journal.pone.0227558. eCollection 2020. PLoS One. 2020. PMID: 31923213 Free PMC article.
-
Polymerase read-through at the first transcription termination site contributes to regulation of borna disease virus gene expression.J Virol. 2008 Oct;82(19):9537-45. doi: 10.1128/JVI.00639-08. Epub 2008 Jul 23. J Virol. 2008. PMID: 18653450 Free PMC article.
References
-
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120(2):183–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

