Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling
- PMID: 18243100
- PMCID: PMC2275919
- DOI: 10.1016/j.cell.2007.12.016
Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling
Abstract
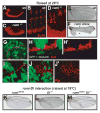
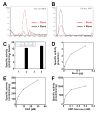
Notch signaling is broadly used to regulate cell-fate decisions. We have identified a gene, rumi, with a temperature-sensitive Notch phenotype. At 28 degrees C-30 degrees C, rumi clones exhibit a full-blown loss of Notch signaling in all tissues tested. However, at 18 degrees C only a mild Notch phenotype is evident. In vivo analyses reveal that the target of Rumi is the extracellular domain of Notch. Notch accumulates intracellularly and at the cell membrane of rumi cells but fails to be properly cleaved, despite normal binding to Delta. Rumi is an endoplasmic reticulum-retained protein with a highly conserved CAP10 domain. Our studies show that Rumi is a protein O-glucosyltransferase, capable of adding glucose to serine residues in Notch EGF repeats with the consensus C1-X-S-X-P-C2 sequence. These data indicate that by O-glucosylating Notch in the ER, Rumi regulates its folding and/or trafficking and allows signaling at the cell membrane.
Figures





Comment in
-
A notch sweeter.Cell. 2008 Jan 25;132(2):177-9. doi: 10.1016/j.cell.2008.01.005. Cell. 2008. PMID: 18243091
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocrine reviews. 2007;28:339–363. - PubMed
-
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Molecular cell. 2000;5:207–216. - PubMed
-
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

