High-resolution mapping and characterization of open chromatin across the genome
- PMID: 18243105
- PMCID: PMC2669738
- DOI: 10.1016/j.cell.2007.12.014
High-resolution mapping and characterization of open chromatin across the genome
Abstract
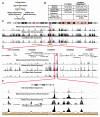
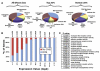
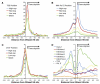
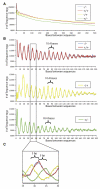
Mapping DNase I hypersensitive (HS) sites is an accurate method of identifying the location of genetic regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions. We employed high-throughput sequencing and whole-genome tiled array strategies to identify DNase I HS sites within human primary CD4+ T cells. Combining these two technologies, we have created a comprehensive and accurate genome-wide open chromatin map. Surprisingly, only 16%-21% of the identified 94,925 DNase I HS sites are found in promoters or first exons of known genes, but nearly half of the most open sites are in these regions. In conjunction with expression, motif, and chromatin immunoprecipitation data, we find evidence of cell-type-specific characteristics, including the ability to identify transcription start sites and locations of different chromatin marks utilized in these cells. In addition, and unexpectedly, our analyses have uncovered detailed features of nucleosome structure.
Figures

References
-
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. - PubMed
-
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the Mamm. Genome. Science. 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

