The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse
- PMID: 18245285
- PMCID: PMC2293198
- DOI: 10.1128/JB.01695-07
The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse
Abstract
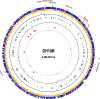
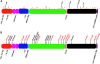
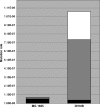
Escherichia coli DH10B was designed for the propagation of large insert DNA library clones. It is used extensively, taking advantage of properties such as high DNA transformation efficiency and maintenance of large plasmids. The strain was constructed by serial genetic recombination steps, but the underlying sequence changes remained unverified. We report the complete genomic sequence of DH10B by using reads accumulated from the bovine sequencing project at Baylor College of Medicine and assembled with DNAStar's SeqMan genome assembler. The DH10B genome is largely colinear with that of the wild-type K-12 strain MG1655, although it is substantially more complex than previously appreciated, allowing DH10B biology to be further explored. The 226 mutated genes in DH10B relative to MG1655 are mostly attributable to the extensive genetic manipulations the strain has undergone. However, we demonstrate that DH10B has a 13.5-fold higher mutation rate than MG1655, resulting from a dramatic increase in insertion sequence (IS) transposition, especially IS150. IS elements appear to have remodeled genome architecture, providing homologous recombination sites for a 113,260-bp tandem duplication and an inversion. DH10B requires leucine for growth on minimal medium due to the deletion of leuLABCD and harbors both the relA1 and spoT1 alleles causing both sensitivity to nutritional downshifts and slightly lower growth rates relative to the wild type. Finally, while the sequence confirms most of the reported alleles, the sequence of deoR is wild type, necessitating reexamination of the assumed basis for the high transformability of DH10B.
Figures



References
-
- Allex, C. F., S. F. Baldwin, J. W. Shavlik, and F. R. Blattner. 1997. Increasing consensus accuracy in DNA fragment assemblies by incorporating fluorescent trace representations. Proc. Int. Conf. Intell. Syst. Mol. Biol. 53-14. - PubMed
-
- Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 21 February 2006, posting date. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. doi:10.1038/msb4100050. - DOI - PMC - PubMed
-
- Bachmann, B. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, DC.
-
- Baev, M. V., D. Baev, A. J. Radek, and J. W. Campbell. 2006. Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of amino acids, peptides, and nucleotides with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71317-322. - PubMed
-
- Beckwith, J. R., and E. R. Signer. 1966. Transposition of the lac region of Escherichia coli. I. Inversion of the lac operon and transduction of lac by phi80. J. Mol. Biol. 19254-265. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

