Low levels of polymorphism in genes that control the activation of defense response in Arabidopsis thaliana
- PMID: 18245336
- PMCID: PMC2323794
- DOI: 10.1534/genetics.107.083279
Low levels of polymorphism in genes that control the activation of defense response in Arabidopsis thaliana
Abstract
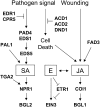
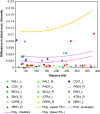
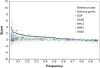
Plants use signaling pathways involving salicylic acid, jasmonic acid, and ethylene to defend against pathogen and herbivore attack. Many defense response genes involved in these signaling pathways have been characterized, but little is known about the selective pressures they experience. A representative set of 27 defense response genes were resequenced in a worldwide set of 96 Arabidopsis thaliana accessions, and patterns of single nucleotide polymorphisms (SNPs) were evaluated in relation to an empirical distribution of SNPs generated from either 876 fragments or 236 fragments with >400 bp coding sequence (this latter set was selected for comparisons with coding sequences) distributed across the genomes of the same set of accessions. Defense response genes have significantly fewer protein variants, display lower levels of nonsynonymous nucleotide diversity, and have fewer nonsynonymous segregating sites. The majority of defense response genes appear to be experiencing purifying selection, given the dearth of protein variation in this set of genes. Eight genes exhibit some evidence of partial selective sweeps or transient balancing selection. These results therefore provide a strong contrast to the high levels of balancing selection exhibited by genes at the upstream positions in these signaling pathways.
Figures

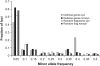

References
-
- Aguadé, M., 2001. Nucleotide sequence variation at two genes of the phenylpropanoid pathway, the FAH1 and F3H genes, in Arabidopsis thaliana. Mol. Biol. Evol. 18 1–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

