Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes
- PMID: 18245380
- PMCID: PMC2542872
- DOI: 10.1073/pnas.0711573105
Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes
Erratum in
- Proc Natl Acad Sci U S A. 2008 May 6;105(18):6787
Abstract
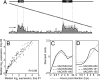
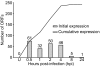
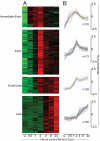
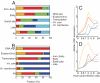
Vaccinia virus is the prototypic orthopoxvirus and was the vaccine used to eradicate smallpox, yet the expression profiles of many of its genes remain unknown. Using a genome tiling array approach, we simultaneously measured the expression levels of all 223 annotated vaccinia virus genes during infection and determined their kinetics. For 95% of these genes, significant transcript levels were detected. Most remarkably, classification of the genes by their expression profiles revealed 35 genes exhibiting immediate-early expression. Although a similar kinetic class has been described for other virus families, to our knowledge, this is the first demonstration of its existence in orthopoxviruses. Despite expression levels higher than for genes in the other three kinetic classes, the functions of more than half of these remain unknown. Additionally, genes within each kinetic class were spatially grouped together in the genome. This genome-wide picture of transcription alters our understanding of how orthopoxviruses regulate gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Poxvirus transcriptome analysis.Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):E62; author reply E63-4. doi: 10.1073/pnas.0804594105. Epub 2008 Oct 1. Proc Natl Acad Sci U S A. 2008. PMID: 18832142 Free PMC article. No abstract available.
References
-
- Pedle S, Cooper RJ. The inhibition of HELA cell RNA synthesis following infection with vaccinia virus. J Gen Virol. 1984;65:1687–1697. - PubMed
-
- Bablanian R, Coppola G, Scribani S, Esteban M. Inhibition of protein synthesis by vaccinia virus. IV. The role of low-molecular-weight viral RNA in the inhibition of protein synthesis. Virology. 1981;112:13–24. - PubMed
-
- Bablanian R, Baxt B, Sonnabend JA, Esteban M. Studies on the mechanisms of vaccinia virus cytopathic effects. II. Early cell rounding is associated with virus polypeptide synthesis. J Gen Virol. 1978;39:403–413. - PubMed
-
- Broyles SS. Vaccinia virus transcription. J Gen Virol. 2003;84:2293–2303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

