Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae
- PMID: 18245831
- PMCID: PMC2278085
- DOI: 10.1534/genetics.107.083535
Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae
Abstract
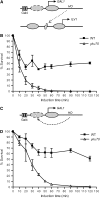
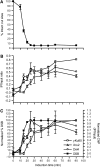
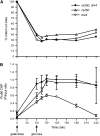
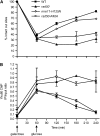
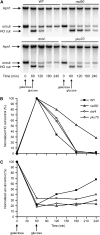
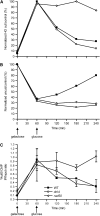
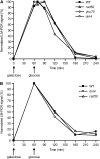
Nonhomologous end joining (NHEJ) is an important DNA double-strand-break (DSB) repair pathway that requires three protein complexes in Saccharomyces cerevisiae: the Ku heterodimer (Yku70-Yku80), MRX (Mre11-Rad50-Xrs2), and DNA ligase IV (Dnl4-Lif1), as well as the ligase-associated protein Nej1. Here we use chromatin immunoprecipitation from yeast to dissect the recruitment and release of these protein complexes at HO-endonuclease-induced DSBs undergoing productive NHEJ. Results revealed that Ku and MRX assembled at a DSB independently and rapidly after DSB formation. Ligase IV appeared at the DSB later than Ku and MRX and in a strongly Ku-dependent manner. Ligase binding was extensive but slightly delayed in rad50 yeast. Ligase IV binding occurred independently of Nej1, but instead promoted loading of Nej1. Interestingly, dissociation of Ku and ligase from unrepaired DSBs depended on the presence of an intact MRX complex and ATP binding by Rad50, suggesting a possible role of MRX in terminating a NHEJ repair phase. This activity correlated with extended DSB resection, but limited degradation of DSB ends occurred even in MRX mutants with persistently bound Ku. These findings reveal the in vivo assembly of the NHEJ repair complex and shed light on the mechanisms controlling DSB repair pathway utilization.
Figures
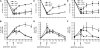

References
-
- Anderson, D. E., K. M. Trujillo, P. Sung and H. P. Erickson, 2001. Structure of the Rad50 × Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 276 37027–37033. - PubMed
-
- Aparicio, O., J. V. Geisberg, E. Sekinger, A. Yang, Z. Moqtaderi et al., 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, pp. 21.3.1–21.3.33 in Current Protocols in Molecular Biology, edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith and K. Struhl. John Wiley & Sons, New York. - PubMed
-
- Boeger, H., J. Griesenbeck, J. S. Strattan and R. D. Kornberg, 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11 1587–1598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

