The glucose transporter (GLUT4) enhancer factor is required for normal wing positioning in Drosophila
- PMID: 18245850
- PMCID: PMC2248331
- DOI: 10.1534/genetics.107.078030
The glucose transporter (GLUT4) enhancer factor is required for normal wing positioning in Drosophila
Abstract
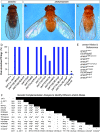
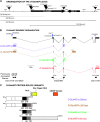
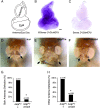
Many of the transcription factors and target genes that pattern the developing adult remain unknown. In the present study, we find that an ortholog of the poorly understood transcription factor, glucose transporter (GLUT4) enhancer factor (Glut4EF, GEF) [also known as the Huntington's disease gene regulatory region-binding protein (HDBP) 1], plays a critical role in specifying normal wing positioning in adult Drosophila. Glut4EF proteins are zinc-finger transcription factors named for their ability to regulate expression of GLUT4 but nothing is known of Glut4EF's in vivo physiological functions. Here, we identify a family of Glut4EF proteins that are well conserved from Drosophila to humans and find that mutations in Drosophila Glut4EF underlie the wing-positioning defects seen in stretch mutants. In addition, our results indicate that previously uncharacterized mutations in Glut4EF are present in at least 11 publicly available fly lines and on the widely used TM3 balancer chromosome. These results indicate that previous observations utilizing these common stocks may be complicated by the presence of Glut4EF mutations. For example, our results indicate that Glut4EF mutations are also present on the same chromosome as two gain-of-function mutations of the homeobox transcription factor Antennapedia (Antp) and underlie defects previously attributed to Antp. In fact, our results support a role for Glut4EF in the modulation of morphogenetic processes mediated by Antp, further highlighting the importance of Glut4EF transcription factors in patterning and morphogenesis.
Figures



Similar articles
-
[Novel miniature-like m42 mutant of Drosophila virilis. Unusual microstructure of wing fibres].Tsitol Genet. 2004 May-Jun;38(3):52-9. Tsitol Genet. 2004. PMID: 15619989 Russian.
-
Activation of the knirps locus links patterning to morphogenesis of the second wing vein in Drosophila.Development. 2003 Jan;130(2):235-48. doi: 10.1242/dev.00207. Development. 2003. PMID: 12466192
-
optomotor-blind suppresses instability at the A/P compartment boundary of the Drosophila wing.Mech Dev. 2008 Mar-Apr;125(3-4):233-46. doi: 10.1016/j.mod.2007.11.006. Epub 2007 Nov 24. Mech Dev. 2008. PMID: 18171611
-
Patterns in evolution: veins of the Drosophila wing.Trends Genet. 2004 Oct;20(10):498-505. doi: 10.1016/j.tig.2004.07.013. Trends Genet. 2004. PMID: 15363904 Review.
-
hedgehog and wing development in Drosophila: a morphogen at work?Bioessays. 2000 May;22(5):460-8. doi: 10.1002/(SICI)1521-1878(200005)22:5<460::AID-BIES8>3.0.CO;2-G. Bioessays. 2000. PMID: 10797486 Review.
Cited by
-
Genome-wide association analysis of tolerance to methylmercury toxicity in Drosophila implicates myogenic and neuromuscular developmental pathways.PLoS One. 2014 Oct 31;9(10):e110375. doi: 10.1371/journal.pone.0110375. eCollection 2014. PLoS One. 2014. PMID: 25360876 Free PMC article.
-
Identification and Characterization of Breakpoints and Mutations on Drosophila melanogaster Balancer Chromosomes.G3 (Bethesda). 2020 Nov 5;10(11):4271-4285. doi: 10.1534/g3.120.401559. G3 (Bethesda). 2020. PMID: 32972999 Free PMC article.
-
Spatial expression of transcription factors in Drosophila embryonic organ development.Genome Biol. 2013 Dec 20;14(12):R140. doi: 10.1186/gb-2013-14-12-r140. Genome Biol. 2013. PMID: 24359758 Free PMC article.
-
Third Chromosome Balancer Inversions Disrupt Protein-Coding Genes and Influence Distal Recombination Events in Drosophila melanogaster.G3 (Bethesda). 2016 Jul 7;6(7):1959-67. doi: 10.1534/g3.116.029330. G3 (Bethesda). 2016. PMID: 27172211 Free PMC article.
-
The Role of the C-Clamp in Wnt-Related Colorectal Cancers.Cancers (Basel). 2016 Aug 3;8(8):74. doi: 10.3390/cancers8080074. Cancers (Basel). 2016. PMID: 27527215 Free PMC article. Review.
References
-
- Arce, L., N. N. Yokoyama and M. L. Waterman, 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25 7492–7504. - PubMed
-
- Atcha, F. A., J. E. Munguia, T. W. Li, K. Hovanes and M. L. Waterman, 2003. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 278 16169–16175. - PubMed
-
- Bate, M., 1993. The mesoderm and its derivatives, pp. 1013–1090 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Bejsovec, A., 2006. Flying at the head of the pack: Wnt biology in Drosophila. Oncogene 25 7442–7449. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

