Genetic evidence for sites of interaction between the Gal3 and Gal80 proteins of the Saccharomyces cerevisiae GAL gene switch
- PMID: 18245852
- PMCID: PMC2248352
- DOI: 10.1534/genetics.107.074799
Genetic evidence for sites of interaction between the Gal3 and Gal80 proteins of the Saccharomyces cerevisiae GAL gene switch
Abstract
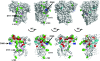
Galactose-activated transcription of the Saccharomyces cerevisiae GAL genes occurs when Gal3 binds the Gal4 inhibitor, Gal80. Noninteracting variants of Gal3 or Gal80 render the GAL genes noninducible. To identify the binding determinants for Gal3's interaction with Gal80 we carried out GAL3-GAL80 intergenic suppression analyses and selected for new GAL3 mutations that impair the Gal3-Gal80 interaction. We show that a GAL3(C)-D368V mutation can suppress the noninducibility due to a GAL80(S-1)-G323R mutation, and a GAL80-M350C mutation can suppress the noninducibility due to a gal3-D111C mutation. A reverse two-hybrid selection for GAL3 mutations that impair the Gal3-Gal80 interaction yielded 12 single-amino-acid substitutions at residues that are predicted to be surface exposed on Gal3. The majority of the affected Gal3 residues localized to a composite surface that includes D111 and a sequence motif containing D368, which has been implicated in interaction with Gal80. The striking colocalization of intergenic suppressor residues and Gal80 nonbinder residues identifies a Gal3 surface that likely interacts with Gal80.
Figures







Similar articles
-
Self-association of the Gal4 inhibitor protein Gal80 is impaired by Gal3: evidence for a new mechanism in the GAL gene switch.Mol Cell Biol. 2013 Sep;33(18):3667-74. doi: 10.1128/MCB.00646-12. Epub 2013 Jul 15. Mol Cell Biol. 2013. PMID: 23858060 Free PMC article.
-
Intragenic suppression of Gal3C interaction with Gal80 in the Saccharomyces cerevisiae GAL gene switch.Genetics. 2006 Jan;172(1):77-87. doi: 10.1534/genetics.105.050807. Epub 2005 Oct 11. Genetics. 2006. PMID: 16219783 Free PMC article.
-
Rapid GAL gene switch of Saccharomyces cerevisiae depends on nuclear Gal3, not nucleocytoplasmic trafficking of Gal3 and Gal80.Genetics. 2011 Nov;189(3):825-36. doi: 10.1534/genetics.111.131839. Epub 2011 Sep 2. Genetics. 2011. PMID: 21890741 Free PMC article.
-
Epigenetics of the yeast galactose genetic switch.J Biosci. 2009 Oct;34(4):513-22. doi: 10.1007/s12038-009-0070-y. J Biosci. 2009. PMID: 19920337 Review.
-
Transcriptional regulation in the yeast GAL gene family: a complex genetic network.FASEB J. 1995 Jun;9(9):777-87. doi: 10.1096/fasebj.9.9.7601342. FASEB J. 1995. PMID: 7601342 Review.
Cited by
-
Self-association of the Gal4 inhibitor protein Gal80 is impaired by Gal3: evidence for a new mechanism in the GAL gene switch.Mol Cell Biol. 2013 Sep;33(18):3667-74. doi: 10.1128/MCB.00646-12. Epub 2013 Jul 15. Mol Cell Biol. 2013. PMID: 23858060 Free PMC article.
-
Mediator acts upstream of the transcriptional activator Gal4.PLoS Biol. 2012;10(3):e1001290. doi: 10.1371/journal.pbio.1001290. Epub 2012 Mar 27. PLoS Biol. 2012. PMID: 22479149 Free PMC article.
-
The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.Genes Dev. 2012 Feb 1;26(3):294-303. doi: 10.1101/gad.182691.111. Genes Dev. 2012. PMID: 22302941 Free PMC article.
-
External control of the GAL network in S. cerevisiae: a view from control theory.PLoS One. 2011 Apr 29;6(4):e19353. doi: 10.1371/journal.pone.0019353. PLoS One. 2011. PMID: 21559408 Free PMC article.
-
Molecular simulation and docking studies of Gal1p and Gal3p proteins in the presence and absence of ligands ATP and galactose: implication for transcriptional activation of GAL genes.J Comput Aided Mol Des. 2012 Jul;26(7):847-64. doi: 10.1007/s10822-012-9579-5. Epub 2012 May 26. J Comput Aided Mol Des. 2012. PMID: 22639079
References
-
- Andreassi, J. L. I., and T. S. Leyh, 2004. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry 43 14594–14601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

