RIC-3: a nicotinic acetylcholine receptor chaperone
- PMID: 18246096
- PMCID: PMC2268041
- DOI: 10.1038/sj.bjp.0707661
RIC-3: a nicotinic acetylcholine receptor chaperone
Abstract
RIC-3 is a transmembrane protein which acts as a molecular chaperone of nicotinic acetylcholine receptors (nAChRs). For some nAChR subtypes (such as homomeric alpha7 neuronal nAChRs), RIC-3 is required for efficient receptor folding, assembly and functional expression. In contrast, for other nAChR subtypes (such as heteromeric alpha4beta2 neuronal nAChRs) there have been reports that RIC-3 can both enhance and reduce levels of functional expression. There is also evidence that RIC-3 can modulate maturation of the closely related 5-hydroxytryptamine (5-HT) receptor (5-HT(3)R). As with heteromeric nAChRs, apparently contradictory results have been reported for the influence of RIC-3 on 5-HT(3)R maturation in different expression systems. Recent evidence indicates that these differences in RIC-3 chaperone activity may be influenced by the host cell, suggesting that other proteins may play an important role in modulating the effects of RIC-3 as a chaperone. RIC-3 was originally identified in the nematode Caenorhabditis elegans as the protein encoded by the gene ric-3 (resistance to inhibitors of cholinesterase) and has subsequently been cloned and characterized from mammalian and insect species. This review provides a brief history of RIC-3; from the identification of the ric-3 gene in C. elegans in 1995 to the more recent demonstration of its activity as a nAChR chaperone.
Figures
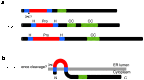
References
-
- Arias HR. Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors. Neurochem Int. 2000;36:595–645. - PubMed
-
- Ben-Ami HC, Yassin L, Farah H, Michaeli A, Eshel M, Treinin M. RIC-3 affects properties and quantity of nicotinic acetylcholine receptors via a mechanism that does not require the coiled-coil domains. J Biol Chem. 2005;280:28053–28060. - PubMed
-
- Bencherif M, Fowler K, Lukas R, Lippiello PM. Mechanisms of up-regulation of neuronal nicotinic acetylcholine receptors in clonal cell lines and primary cultures of fetal rat brain. J Pharmacol Exp Ther. 1995;275:987–994. - PubMed
-
- Benwell MEM, Balfour DJK, Anderson JM. Evidence that tobacco smoking increases the density of (−)-[3H]nicotine binding sites in human brain. J Neurochem. 1988;50:1243–1247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

