A calculated response: control of inflammation by the innate immune system
- PMID: 18246191
- PMCID: PMC2214713
- DOI: 10.1172/JCI34431
A calculated response: control of inflammation by the innate immune system
Abstract
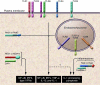
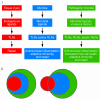
Inflammation is a rapid yet coordinated response that can lead to the destruction of microbes and host tissue. Triggers capable of inducing an inflammatory response include tissue damage and infection by pathogenic and nonpathogenic microbes. Each of these triggers represents a qualitatively distinct stress to the host immune system, yet our understanding of whether they are interpreted as such remains poor. Accumulating evidence suggests that recognition of these distinct stimuli converges on many of the same receptors of the innate immune system. Here I provide an overview of these innate receptors and suggest that the innate immune system can interpret the context of an inflammatory trigger and direct inflammation accordingly.
Figures

References
-
- Kumar, V., Abbas, A.K., Fausto, N., and Mitchell, M. 2007. Robbins basic pathology. W.B. Saunders. Philadelphia, Pennsylvania, USA. 960 pp.
-
- Nathan C. Points of control in inflammation. . Nature. 2002;420:846–852. - PubMed
-
- Segal B.H., Leto T.L., Gallin J.I., Malech H.L., Holland S.M. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine. 2000;79:170–200. - PubMed
-
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. - PubMed
-
- Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

