The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis
- PMID: 18248093
- PMCID: PMC2222957
- DOI: 10.1371/journal.ppat.0040020
The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis
Abstract
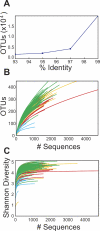
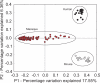
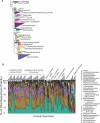
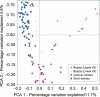
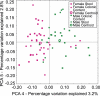
The vertebrate gut harbors a vast community of bacterial mutualists, the composition of which is modulated by the host immune system. Many gastrointestinal (GI) diseases are expected to be associated with disruptions of host-bacterial interactions, but relatively few comprehensive studies have been reported. We have used the rhesus macaque model to investigate forces shaping GI bacterial communities. We used DNA bar coding and pyrosequencing to characterize 141,000 sequences of 16S rRNA genes obtained from 100 uncultured GI bacterial samples, allowing quantitative analysis of community composition in health and disease. Microbial communities of macaques were distinct from those of mice and humans in both abundance and types of taxa present. The macaque communities differed among samples from intestinal mucosa, colonic contents, and stool, paralleling studies of humans. Communities also differed among animals, over time within individual animals, and between males and females. To investigate changes associated with disease, samples of colonic contents taken at necropsy were compared between healthy animals and animals with colitis and undergoing antibiotic therapy. Communities from diseased and healthy animals also differed significantly in composition. This work provides comprehensive data and improved methods for studying the role of commensal microbiota in macaque models of GI diseases and provides a model for the large-scale screening of the human gut microbiome.
Conflict of interest statement
Figures

References
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008759/GM/NIGMS NIH HHS/United States
- P51 RR000164/RR/NCRR NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- RR00164/RR/NCRR NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- AI045008/AI/NIAID NIH HHS/United States
- T32 GM065103/GM/NIGMS NIH HHS/United States
- S10 OD012300/OD/NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- GM08759/GM/NIGMS NIH HHS/United States
- G20 RR016930/RR/NCRR NIH HHS/United States
- RR016930/RR/NCRR NIH HHS/United States
- P01DK078669/DK/NIDDK NIH HHS/United States
- T32GM065103/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

