Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules
- PMID: 18248097
- PMCID: PMC2222925
- DOI: 10.1371/journal.pgen.0040014
Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules
Abstract
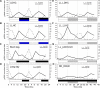
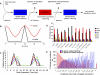
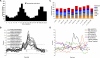
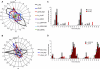
Correct daily phasing of transcription confers an adaptive advantage to almost all organisms, including higher plants. In this study, we describe a hypothesis-driven network discovery pipeline that identifies biologically relevant patterns in genome-scale data. To demonstrate its utility, we analyzed a comprehensive matrix of time courses interrogating the nuclear transcriptome of Arabidopsis thaliana plants grown under different thermocycles, photocycles, and circadian conditions. We show that 89% of Arabidopsis transcripts cycle in at least one condition and that most genes have peak expression at a particular time of day, which shifts depending on the environment. Thermocycles alone can drive at least half of all transcripts critical for synchronizing internal processes such as cell cycle and protein synthesis. We identified at least three distinct transcription modules controlling phase-specific expression, including a new midnight specific module, PBX/TBX/SBX. We validated the network discovery pipeline, as well as the midnight specific module, by demonstrating that the PBX element was sufficient to drive diurnal and circadian condition-dependent expression. Moreover, we show that the three transcription modules are conserved across Arabidopsis, poplar, and rice. These results confirm the complex interplay between thermocycles, photocycles, and the circadian clock on the daily transcription program, and provide a comprehensive view of the conserved genomic targets for a transcriptional network key to successful adaptation.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

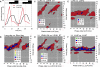
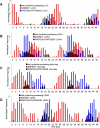
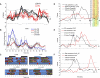
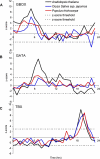
Similar articles
-
Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules.PLoS One. 2011;6(6):e16907. doi: 10.1371/journal.pone.0016907. Epub 2011 Jun 9. PLoS One. 2011. PMID: 21694767 Free PMC article.
-
A morning-specific phytohormone gene expression program underlying rhythmic plant growth.PLoS Biol. 2008 Sep 16;6(9):e225. doi: 10.1371/journal.pbio.0060225. PLoS Biol. 2008. PMID: 18798691 Free PMC article.
-
The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis.Cold Spring Harb Symp Quant Biol. 2007;72:353-63. doi: 10.1101/sqb.2007.72.006. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419293
-
The Transcriptional Network in the Arabidopsis Circadian Clock System.Genes (Basel). 2020 Oct 29;11(11):1284. doi: 10.3390/genes11111284. Genes (Basel). 2020. PMID: 33138078 Free PMC article. Review.
-
Arabidopsis transcription factors and the regulation of flowering time: a genomic perspective.Curr Issues Mol Biol. 2002 Jul;4(3):77-91. Curr Issues Mol Biol. 2002. PMID: 12074197 Review.
Cited by
-
Genome-wide profiling of 24 hr diel rhythmicity in the water flea, Daphnia pulex: network analysis reveals rhythmic gene expression and enhances functional gene annotation.BMC Genomics. 2016 Aug 18;17:653. doi: 10.1186/s12864-016-2998-2. BMC Genomics. 2016. PMID: 27538446 Free PMC article.
-
Crosstalk between the circadian clock and innate immunity in Arabidopsis.PLoS Pathog. 2013;9(6):e1003370. doi: 10.1371/journal.ppat.1003370. Epub 2013 Jun 6. PLoS Pathog. 2013. PMID: 23754942 Free PMC article.
-
Genomic Analysis Reveals Contrasting PIFq Contribution to Diurnal Rhythmic Gene Expression in PIF-Induced and -Repressed Genes.Front Plant Sci. 2016 Jul 4;7:962. doi: 10.3389/fpls.2016.00962. eCollection 2016. Front Plant Sci. 2016. PMID: 27458465 Free PMC article.
-
Unlocking Nature's Rhythms: Insights into Secondary Metabolite Modulation by the Circadian Clock.Int J Mol Sci. 2024 Jul 3;25(13):7308. doi: 10.3390/ijms25137308. Int J Mol Sci. 2024. PMID: 39000414 Free PMC article. Review.
-
Physiological genomics of response to soil drying in diverse Arabidopsis accessions.Plant Cell. 2012 Mar;24(3):893-914. doi: 10.1105/tpc.112.096180. Epub 2012 Mar 9. Plant Cell. 2012. PMID: 22408074 Free PMC article.
References
-
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. - PubMed
-
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. - PubMed
-
- Woelfle M, Ouyang Y, Phanvijhitsiri K, Johnson C. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. - PubMed
-
- Wijnen H, Young M. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. - PubMed
-
- Lahiri K, Vallone D, Gondi SB, Santoriello C, Dickmeis T, et al. Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol. 2005;3:e351. doi: 10.1371/journal.pbio.0030351. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

