Probing the mechanism of recognition of ssDNA by the Cdc13-DBD
- PMID: 18250086
- PMCID: PMC2275150
- DOI: 10.1093/nar/gkn017
Probing the mechanism of recognition of ssDNA by the Cdc13-DBD
Abstract
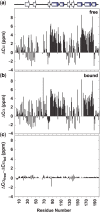
The Saccharomyces cerevisiae protein Cdc13 tightly and specifically binds the conserved G-rich single-stranded overhang at telomeres and plays an essential role in telomere end-protection and length regulation. The 200 residue DNA-binding domain of Cdc13 (Cdc13-DBD) binds an 11mer single-stranded representative of the yeast telomeric sequence [Tel11, d(GTGTGGGTGTG)] with a 3 pM affinity and specificity for three bases (underlined) at the 5' end. The structure of the Cdc13-DBD bound to Tel11 revealed a large, predominantly aromatic protein interface with several unusual features. The DNA adopts an irregular, extended structure, and the binding interface includes a long ( approximately 30 amino acids) structured loop between strands beta2-beta3 (L(2-3)) of an OB-fold. To investigate the mechanism of ssDNA binding, we studied the free and bound states of Cdc13-DBD using NMR spectroscopy. Chemical shift changes indicate that the basic topology of the domain, including L(2-3), is essentially intact in the free state. Changes in slow and intermediate time scale dynamics, however, occur in L(2-3), while conformational changes distant from the DNA interface suggest an induced fit mechanism for binding in the 'hot spot' for binding affinity and specificity. These data point to an overall binding mechanism well adapted to the heterogeneous nature of yeast telomeres.
Figures



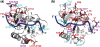
Similar articles
-
Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13.J Mol Biol. 2004 Apr 23;338(2):241-55. doi: 10.1016/j.jmb.2004.01.063. J Mol Biol. 2004. PMID: 15066429
-
Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13.Biochemistry. 2003 Apr 8;42(13):3751-8. doi: 10.1021/bi027047c. Biochemistry. 2003. PMID: 12667066
-
The tenacious recognition of yeast telomere sequence by Cdc13 is fully exerted by a single OB-fold domain.Nucleic Acids Res. 2014 Jan;42(1):475-84. doi: 10.1093/nar/gkt843. Epub 2013 Sep 20. Nucleic Acids Res. 2014. PMID: 24057216 Free PMC article.
-
Themes in ssDNA recognition by telomere-end protection proteins.Trends Biochem Sci. 2006 Sep;31(9):516-25. doi: 10.1016/j.tibs.2006.07.004. Epub 2006 Aug 4. Trends Biochem Sci. 2006. PMID: 16890443 Review.
-
Structural anatomy of telomere OB proteins.Crit Rev Biochem Mol Biol. 2011 Oct;46(5):409-35. doi: 10.3109/10409238.2011.609295. Crit Rev Biochem Mol Biol. 2011. PMID: 21950380 Free PMC article. Review.
Cited by
-
Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p.Nucleic Acids Res. 2009 Feb;37(2):354-67. doi: 10.1093/nar/gkn830. Epub 2008 Nov 29. Nucleic Acids Res. 2009. PMID: 19043074 Free PMC article.
-
Cdc13 exhibits dynamic DNA strand exchange in the presence of telomeric DNA.Nucleic Acids Res. 2024 Jun 24;52(11):6317-6332. doi: 10.1093/nar/gkae265. Nucleic Acids Res. 2024. PMID: 38613387 Free PMC article.
-
Dynamic peptides of human TPP1 fulfill diverse functions in telomere maintenance.Nucleic Acids Res. 2016 Dec 1;44(21):10467-10479. doi: 10.1093/nar/gkw846. Epub 2016 Sep 20. Nucleic Acids Res. 2016. PMID: 27655633 Free PMC article.
-
Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities.Pathogens. 2021 Jul 30;10(8):967. doi: 10.3390/pathogens10080967. Pathogens. 2021. PMID: 34451431 Free PMC article. Review.
-
The mechanisms of K. lactis Cdc13 in telomere DNA-binding and telomerase regulation.DNA Repair (Amst). 2018 Jan;61:37-45. doi: 10.1016/j.dnarep.2017.11.007. Epub 2017 Nov 28. DNA Repair (Amst). 2018. PMID: 29197718 Free PMC article.
References
-
- Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem. Sci. 2006;31:516–525. - PubMed
-
- Schuermann JP, Prewitt SP, Davies C, Deutscher SL, Tanner JJ. Evidence for structural plasticity of heavy chain complementarity-determining region 3 in antibody-ssDNA recognition. J. Mol. Biol. 2005;347:965–978. - PubMed
-
- Lei M, Podell E, Cech TR. Structure of human Pot1 bound to telomeric DNA single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. - PubMed
-
- Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the crystal structure of the Pot1 (Protection of Telomeres)-ssDNA complex. Nature. 2003;426:198–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

