Genome-wide approaches to studying chromatin modifications
- PMID: 18250624
- PMCID: PMC10882563
- DOI: 10.1038/nrg2270
Genome-wide approaches to studying chromatin modifications
Abstract
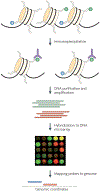
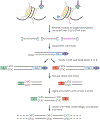
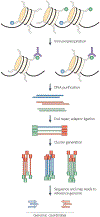
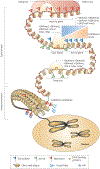
Over two metres of DNA is packaged into each nucleus in the human body in a manner that still allows for gene regulation. This remarkable feat is accomplished by the wrapping of DNA around histone proteins in repeating units of nucleosomes to form a structure known as chromatin. This chromatin structure is subject to various modifications that have profound influences on gene expression. Recently developed techniques to study chromatin modifications at a genome-wide scale are now allowing researchers to probe the complex components that make up epigenomes. Here we review genome-wide approaches to studying epigenomic structure and the exciting findings that have been obtained using these technologies.
Figures

References
-
- Goldberg AD, Allis CD & Bernstein E Epigenetics: a landscape takes shape. Cell 128, 635–638 (2007). - PubMed
-
- Ptashne M On the use of the word ‘epigenetic’. Curr. Biol 17, R233–R236 (2007). - PubMed
-
- Bird A Perceptions of epigenetics. Nature 447, 396–398 (2007). - PubMed
-
- Bernstein BE, Meissner A & Lander ES The mammalian epigenome. Cell 128, 669–681 (2007). - PubMed
-
- Orlando V & Paro R Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75, 1187–1198 (1993). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

