Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping
- PMID: 18251999
- PMCID: PMC2259380
- DOI: 10.1186/1471-2156-9-14
Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping
Abstract
Background: Genetic isolates such as the Ashkenazi Jews (AJ) potentially offer advantages in mapping novel loci in whole genome disease association studies. To analyze patterns of genetic variation in AJ, genotypes of 101 healthy individuals were determined using the Affymetrix EAv3 500 K SNP array and compared to 60 CEPH-derived HapMap (CEU) individuals. 435,632 SNPs overlapped and met annotation criteria in the two groups.
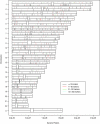
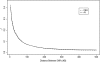
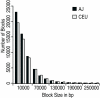
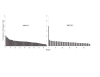
Results: A small but significant global difference in allele frequencies between AJ and CEU was demonstrated by a mean FST of 0.009 (P < 0.001); large regions that differed were found on chromosomes 2 and 6. Haplotype blocks inferred from pairwise linkage disequilibrium (LD) statistics (Haploview) as well as by expectation-maximization haplotype phase inference (HAP) showed a greater number of haplotype blocks in AJ compared to CEU by Haploview (50,397 vs. 44,169) or by HAP (59,269 vs. 54,457). Average haplotype blocks were smaller in AJ compared to CEU (e.g., 36.8 kb vs. 40.5 kb HAP). Analysis of global patterns of local LD decay for closely-spaced SNPs in CEU demonstrated more LD, while for SNPs further apart, LD was slightly greater in the AJ. A likelihood ratio approach showed that runs of homozygous SNPs were approximately 20% longer in AJ. A principal components analysis was sufficient to completely resolve the CEU from the AJ.
Conclusion: LD in the AJ versus was lower than expected by some measures and higher by others. Any putative advantage in whole genome association mapping using the AJ population will be highly dependent on regional LD structure.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

