A unified association analysis approach for family and unrelated samples correcting for stratification
- PMID: 18252216
- PMCID: PMC2427300
- DOI: 10.1016/j.ajhg.2007.10.009
A unified association analysis approach for family and unrelated samples correcting for stratification
Abstract
There are two common designs for association mapping of complex diseases: case-control and family-based designs. A case-control sample is more powerful to detect genetic effects than a family-based sample that contains the same numbers of affected and unaffected persons, although additional markers may be required to control for spurious association. When family and unrelated samples are available, statistical analyses are often performed in the family and unrelated samples separately, conditioning on parental information for the former, thus resulting in reduced power. In this report, we propose a unified approach that can incorporate both family and case-control samples and, provided the additional markers are available, at the same time corrects for population stratification. We apply the principal components of a marker matrix to adjust for the effect of population stratification. This unified approach makes it unnecessary to perform a conditional analysis of the family data and is more powerful than the separate analyses of unrelated and family samples, or a meta-analysis performed by combining the results of the usual separate analyses. This property is demonstrated in both a variety of simulation models and empirical data. The proposed approach can be equally applied to the analysis of both qualitative and quantitative traits.
Figures
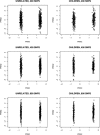



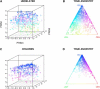
References
-
- Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. - PubMed
-
- Risch N.J. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. - PubMed
-
- Lander E.S., Schork N.J. Genetic dissection of complex traits. Science. 1994;265:2037–2048. - PubMed
-
- Marchini J., Cardon L.R., Phillips M.S., Donnelly P. The effects of human population structure on large genetic association studies. Nat. Genet. 2004;36:512–517. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

