Identification of the Otopetrin Domain, a conserved domain in vertebrate otopetrins and invertebrate otopetrin-like family members
- PMID: 18254951
- PMCID: PMC2268672
- DOI: 10.1186/1471-2148-8-41
Identification of the Otopetrin Domain, a conserved domain in vertebrate otopetrins and invertebrate otopetrin-like family members
Abstract
Background: Otopetrin 1 (Otop1) encodes a multi-transmembrane domain protein with no homology to known transporters, channels, exchangers, or receptors. Otop1 is necessary for the formation of otoconia and otoliths, calcium carbonate biominerals within the inner ear of mammals and teleost fish that are required for the detection of linear acceleration and gravity. Vertebrate Otop1 and its paralogues Otop2 and Otop3 define a new gene family with homology to the invertebrate Domain of Unknown Function 270 genes (DUF270; pfam03189).
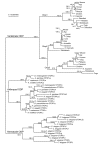
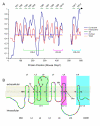
Results: Multi-species comparison of the predicted primary sequences and predicted secondary structures of 62 vertebrate otopetrin, and arthropod and nematode DUF270 proteins, has established that the genes encoding these proteins constitute a single family that we renamed the Otopetrin Domain Protein (ODP) gene family. Signature features of ODP proteins are three "Otopetrin Domains" that are highly conserved between vertebrates, arthropods and nematodes, and a highly constrained predicted loop structure.
Conclusion: Our studies suggest a refined topologic model for ODP insertion into the lipid bilayer of 12 transmembrane domains, and highlight conserved amino-acid residues that will aid in the biochemical examination of ODP family function. The high degree of sequence and structural similarity of the ODP proteins may suggest a conserved role in the intracellular trafficking of calcium and the formation of biominerals.
Figures

References
-
- Oas JG. Benign paroxysmal positional vertigo: a clinician's perspective. Ann N Y Acad Sci. 2001;942:201–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

