A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish
- PMID: 18254971
- PMCID: PMC2268669
- DOI: 10.1186/1471-2148-8-42
A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish
Abstract
Background: A large multigene family of NOD-like receptor (NLR) molecules have been described in mammals and implicated in immunity and apoptosis. Little information, however, exists concerning this gene family in non-mammalian taxa. This current study, therefore, provides an in-depth investigation of this gene family in lower vertebrates including extensive phylogenetic comparison of zebrafish NLRs with orthologs in tetrapods, and analysis of their tissue-specific expression.
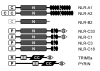
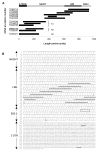
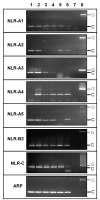
Results: Three distinct NLR subfamilies were identified by mining genome databases of various non-mammalian vertebrates; the first subfamily (NLR-A) resembles mammalian NODs, the second (NLR-B) resembles mammalian NALPs, while the third (NLR-C) appears to be unique to teleost fish. In zebrafish, NLR-A and NLR-B subfamilies contain five and six genes respectively. The third subfamily is large, containing several hundred NLR-C genes, many of which are predicted to encode a C-terminal B30.2 domain. This subfamily most likely evolved from a NOD3-like molecule. Gene predictions for zebrafish NLRs were verified using sequence derived from ESTs or direct sequencing of cDNA. Reverse-transcriptase (RT)-PCR analysis confirmed expression of representative genes from each subfamily in selected tissues.
Conclusion: Our findings confirm the presence of multiple NLR gene orthologs, which form a large multigene family in teleostei. Although the functional significance of the three major NLR subfamilies is unclear, we speculate that conservation and abundance of NLR molecules in all teleostei genomes, reflects an essential role in cellular control, apoptosis or immunity throughout bony fish.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

