RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses
- PMID: 18256230
- PMCID: PMC2292964
- DOI: 10.1128/JCM.02294-07
RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses
Abstract
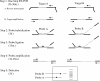
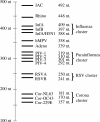
Broad-spectrum analysis for pathogens in patients with respiratory tract infections is becoming more relevant as the number of potential infectious agents is still increasing. Here we describe the new multiparameter RespiFinder assay, which is based on the multiplex ligation-dependent probe amplification (MLPA) technology. This assay detects 15 respiratory viruses in one reaction. The MLPA reaction is preceded by a preamplification step which ensures the detection of both RNA and DNA viruses with the same specificity and sensitivity as individual monoplex real-time reverse transcription-PCRs. The RespiFinder assay was validated with 144 clinical samples, and the results of the assay were compared to those of cell culture and a respiratory syncytial virus (RSV)-specific immunochromatography assay (ICA). Compared to the cell culture results, the RespiFinder assay showed specificities and sensitivities of 98.2% and 100%, respectively, for adenovirus; 96.4% and 100%, respectively, for human metapneumovirus; 98.2% and 100%, respectively, for influenza A virus (InfA); 99.1% and 100%, respectively, for parainfluenza virus type 1 (PIV-1); 99.1% and 80%, respectively, for PIV-3; 90.1% and 100%, respectively, for rhinovirus; and 94.6% and 100%, respectively, for RSV. Compared to the results of the RSV-specific ICA, the RespiFinder assay gave a specificity and a sensitivity of 82.4% and 80%, respectively. PIV-2, PIV-4, influenza B virus, InfA H5N1, and coronavirus 229E were not detected in the clinical specimens tested. The use of the RespiFinder assay resulted in an increase in the diagnostic yield compared to that obtained by cell culture (diagnostic yields, 60% and 35.5%, respectively). In conclusion, the RespiFinder assay provides a user-friendly and high-throughput tool for the simultaneous detection of 15 respiratory viruses with excellent overall performance statistics.
Figures


Similar articles
-
Comparison of three multiplex PCR assays for the detection of respiratory viral infections: evaluation of xTAG respiratory virus panel fast assay, RespiFinder 19 assay and RespiFinder SMART 22 assay.BMC Infect Dis. 2012 Jul 24;12:163. doi: 10.1186/1471-2334-12-163. BMC Infect Dis. 2012. PMID: 22828244 Free PMC article.
-
Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections.J Clin Microbiol. 2007 Jul;45(7):2105-9. doi: 10.1128/JCM.00210-07. Epub 2007 May 16. J Clin Microbiol. 2007. PMID: 17507510 Free PMC article.
-
Detection and typing by molecular techniques of respiratory viruses in children hospitalized for acute respiratory infection in Rome, Italy.J Med Virol. 2007 Apr;79(4):463-8. doi: 10.1002/jmv.20832. J Med Virol. 2007. PMID: 17311326 Free PMC article.
-
The diagnosis of viral respiratory disease in older adults.Clin Infect Dis. 2010 Mar 1;50(5):747-51. doi: 10.1086/650486. Clin Infect Dis. 2010. PMID: 20121411 Free PMC article. Review.
-
[Procedures of viral identification in respiratory infections].Arch Bronconeumol. 1995 Nov;31(9):470-80. doi: 10.1016/s0300-2896(15)30868-1. Arch Bronconeumol. 1995. PMID: 8520820 Free PMC article. Review. Spanish. No abstract available.
Cited by
-
Practical Prediction of Ten Common Streptococcus pneumoniae Serotypes/Serogroups in One PCR Reaction by Multiplex Ligation-Dependent Probe Amplification and Melting Curve (MLPA-MC) Assay in Shenzhen, China.PLoS One. 2015 Jul 7;10(7):e0130664. doi: 10.1371/journal.pone.0130664. eCollection 2015. PLoS One. 2015. PMID: 26151828 Free PMC article.
-
Comparison of three multiplex PCR assays for the detection of respiratory viral infections: evaluation of xTAG respiratory virus panel fast assay, RespiFinder 19 assay and RespiFinder SMART 22 assay.BMC Infect Dis. 2012 Jul 24;12:163. doi: 10.1186/1471-2334-12-163. BMC Infect Dis. 2012. PMID: 22828244 Free PMC article.
-
Human Metapneumovirus: lessons learned over the first decade.Clin Microbiol Rev. 2011 Oct;24(4):734-54. doi: 10.1128/CMR.00015-11. Clin Microbiol Rev. 2011. PMID: 21976607 Free PMC article. Review.
-
A multiplex oligonucleotide ligation-PCR as a complementary tool for subtyping of Salmonella Typhimurium.Appl Microbiol Biotechnol. 2015 Oct;99(19):8137-49. doi: 10.1007/s00253-015-6831-7. Epub 2015 Jul 25. Appl Microbiol Biotechnol. 2015. PMID: 26205523 Free PMC article.
-
Molecular detection of respiratory viruses in immunocopromised ICU patients: incidence and meaning.Respir Med. 2012 Aug;106(8):1184-91. doi: 10.1016/j.rmed.2012.05.001. Epub 2012 May 29. Respir Med. 2012. PMID: 22647492 Free PMC article.
References
-
- Beld, M., R. Minnaar, J. Weel, C. Sol, M. Damen, H. van der Avoort, P. Wertheim-van Dillen, A. van Breda, and R. Boom. 2004. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J. Clin. Microbiol. 423059-3064. - PMC - PubMed
-
- Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

