WGAViewer: software for genomic annotation of whole genome association studies
- PMID: 18256235
- PMCID: PMC2279251
- DOI: 10.1101/gr.071571.107
WGAViewer: software for genomic annotation of whole genome association studies
Abstract
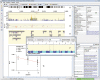
To meet the immediate need for a framework of post-whole genome association (WGA) annotation, we have developed WGAViewer, a suite of JAVA software tools that provides a user-friendly interface to automatically annotate, visualize, and interpret the set of P-values emerging from a WGA study. Most valuably, it can be used to highlight possible functional mechanisms in an automatic manner, for example, by directly or indirectly implicating a polymorphism with an apparent link to gene expression, and help to generate hypotheses concerning the possible biological bases of observed associations. The easily interpretable diagrams can then be used to identify the associations that seem most likely to be biologically relevant, and to select genomic regions that may need to be resequenced in a search for candidate causal variants. In this report, we used our recently completed study on host control of HIV-1 viral load during the asymptomatic set point period as an illustration for the heuristic annotation of this software and its contributive role in a successful WGA project.
Figures
References
-
- Cavalleri G.L., Weale M.E., Shianna K.V., Singh R., Lynch J.M., Grinton B., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Weale M.E., Shianna K.V., Singh R., Lynch J.M., Grinton B., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Shianna K.V., Singh R., Lynch J.M., Grinton B., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Singh R., Lynch J.M., Grinton B., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Lynch J.M., Grinton B., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Grinton B., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Szoeke C., Murphy K., Kinirons P., O’Rourke D., Murphy K., Kinirons P., O’Rourke D., Kinirons P., O’Rourke D., O’Rourke D., et al. Multicentre search for genetic susceptibility loci in sporadic epilepsy syndrome and seizure types: A case-control study. Lancet Neurol. 2007;6:970–980. - PubMed
-
- Fellay J., Shianna K.V., Ge D., Colombo S., Ledergerber B., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., Shianna K.V., Ge D., Colombo S., Ledergerber B., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., Ge D., Colombo S., Ledergerber B., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., Colombo S., Ledergerber B., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., Ledergerber B., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., Zhang K., Gumbs C., Castagna A., Cossarizza A., Gumbs C., Castagna A., Cossarizza A., Castagna A., Cossarizza A., Cossarizza A., et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. - PMC - PubMed
-
- Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Clarke L., Coates G., Cunningham F., Cutts T., Coates G., Cunningham F., Cutts T., Cunningham F., Cutts T., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. - PMC - PubMed
-
- Kasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E., Hammond M., Rocca-Serra P., Cox T., Birney E., Rocca-Serra P., Cox T., Birney E., Cox T., Birney E., Birney E. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
