A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging
- PMID: 18256261
- PMCID: PMC2902235
- DOI: 10.1523/JNEUROSCI.4098-07.2008
A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging
Abstract
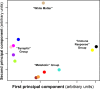
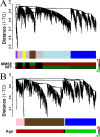
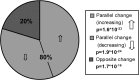
Alzheimer's disease (AD) is a debilitating neurodegenerative disorder affecting millions of elderly individuals worldwide. Advances in the genetics of AD have led to new levels of understanding and treatment opportunities. Here, we used a systems biology approach based on weighted gene coexpression network analysis to determine transcriptional networks in AD. This method permits a higher order depiction of gene expression relationships and identifies modules of coexpressed genes that are functionally related, rather than producing massive gene lists. Using this framework, we characterized the transcriptional network in AD, identifying 12 distinct modules related to synaptic and metabolic processes, immune response, and white matter, nine of which were related to disease progression. We further examined the association of gene expression changes with progression of AD and normal aging, and were able to compare functional modules of genes defined in both conditions. Two biologically relevant modules were conserved between AD and aging, one related to mitochondrial processes such as energy metabolism, and the other related to synaptic plasticity. We also identified several genes that were central, or hub, genes in both aging and AD, including the highly abundant signaling molecule 14.3.3 zeta (YWHAZ), whose role in AD and aging is uncharacterized. Finally, we found that presenilin 1 (PSEN1) is highly coexpressed with canonical myelin proteins, suggesting a role for PSEN1 in aspects of glial-neuronal interactions related to neurodegenerative processes.
Figures




References
-
- Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. - PubMed
-
- Arendt T. Alzheimer's disease as a loss of differentiation control in a subset of neurons that retain immature features in the adult brain. Neurobiol Aging. 2000;21:783–796. - PubMed
-
- Arendt T. Neurodegeneration and plasticity. Int J Dev Neurosci. 2004;22:507–514. - PubMed
-
- Baldi P, Long A. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. - PubMed
-
- Barabasi A, Oltvai Z. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
